
Combined transcriptomic and lipidomic analysis of D-4F ameliorating bleomycin-induced pulmonary fibrosis
Introduction
Idiopathic pulmonary fibrosis (IPF) is a progressive lung disease that leads to respiratory failure. It is characterized by the excessive deposition of the extracellular matrix (ECM) in the pulmonary interstitium (1). fibroblast proliferation and deposition of the ECM are the major pathological changes of IPF (2). Epithelial-to-mesenchymal transition (EMT) describes the process by which epithelial cells transition into mesenchymal cells (3,4). Many factors can induce EMT, including the transforming growth factor β1 (TGF-β1), which is considered the major inducer of EMT (4). Research has shown that the level of apolipoprotein A-1 (ApoA-1) is decreased in IPF patients, and aopa1 can ameliorate BLM-induced lung injury (5).
ApoA-1 is the major component of high-density lipoproteins (HDLs) and plays a key role in regulating normal lipid homeostasis by participating in reverse cholesterol transport (the process by which excess cholesterol in the periphery is transported to the liver and excreted from the body) (6). ApoA‐1 is expressed in alveolar epithelial cells (AECs) and has anti‐inflammatory effects in various lung diseases (7). The overexpression of ApoA-1 in experimental silicosis has been shown to decrease silica-induced lung inflammation and fibrotic nodule formation (8). Thus, ApoA-1 is a potential target for IPF.
D-4F (Ac-DWFKAFYDKVAEKFKEAF-NH2) is an 18-amino acid mimetic peptide of ApoA-1 (9). It contains a class A amphipathic helix with a polar and a non-polar face that binds lipids (10,11). It does not have a homologous sequence with ApoA-1 (10,11). D-4F was initially reported to be a drug treatment of the disease in the cardiovascular system; however, recently, it has been reported to be a drug treatment of the disease in the respiratory system that notably occurs in macrophages (12) and human AECs (12). It was hypothesized that D-4F ameliorates the IPF model in vivo. We conducted an untargeted lipidomic and transcriptomic analysis to investigate the effects of D-4F on the IPF model in vivo. We sought to identify a crucial target and explain the mechanism of D-4F in the IPF model.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3777) (13,14).
Methods
Animals
C57BL/6 male mice were purchased from SPF Biotechnology Co., Ltd. (Beijing, China). This study began when all the mice were 8-weeks-old. Under specific pathogen free (SPF) conditions, the mice were fed an ad-libitum diet of laboratory pellet chow and tap water, and were kept on a 12-hour light/dark cycle at the Animal Centre of Shandong University. tyExperiments were performed under a project license (No.: 21109) granted by Laboratory Animal Ethical and Welfare Committee of Shandong University Cheeloo College of Medicine, in compliance with Shandong University Cheeloo College of Medicine guidelines for the care and use of animals. A protocol was prepared before the study without registration.
Induction of lung fibrosis
Bleomycin hydrochloride was purchased from Hisun Pfizer Pharmaceuticals Co., Ltd. (Zhejiang, China). D-4F (Ac-DWFKAFYDKVAEKFKEAF-NH2) was synthesized by Cloud-Clone Corp (purity >98%, Wuhan, China). Bleomycin and D-4F were dissolved in phosphate-buffered saline (PBS). Because of the minimum sample size of untargeted lipidomic analysis and the mortality of Bleomycin-induced IPF model, 36 C57BL/6 mice enter into this experiment. There were 3 groups in our study and there were 12 mice in each group. All C57BL/6 mice were randomly assigned to 3 groups: the Blank Group (BLK Group), Bleomycin Treatment Group (Model Group), and the D-4F Interference Group (Inter Group).
The way of induction of lung fibrosis is as the article described (15). To induce pulmonary fibrosis, the mice were anesthetized with pentobarbital sodium salt and given a single intratracheal instillation of 60 μL (2 mg/kg) or PBS as a control. After bleomycin or PBS instillation, D-4F (3 mg/kg) was intraperitoneally injected once a day for 4 weeks in the Inter Group. All the mice were anesthetized 4 weeks later. Blood and lung tissue samples were taken. The plasma samples were frozen immediately and stored at −80 °C. The lungs were perfused with PBS to remove blood, and portions of the left lungs were fixed in 4% paraformaldehyde.
Histopathology
The portions of the left lungs underwent hematoxylin and eosin (H&E), Masson’s trichrome (M-T), and immunohistochemical staining. The alpha smooth muscle actin (α-SMA) antibody (Abcam Cat# ab124964, RRID:AB_11129103) was purchased from Abcam (Cambridge, UK). The Ashcroft scale was used to evaluate lung fibrosis (16). The density of the M-T and immunohistochemistry staining of α-SMA were scored using an image J analysis program.
TGF-β1 measurement
TGF-β1 was measured using enzyme-linked immunoassay (ELISA) kits purchased from Beyotime Biotechnology (Shanghai, China).
Untargeted lipidomic analysis
There were 9 samples per group. The samples were analyzed as described previously (17-19). Detailed step-by-step descriptions are provided in the Appendix 1. In brief, after lipid extraction, an untargeted lipidomic analysis, ultra-high performance liquid chromatography (UHPLC)-tandem mass spectrometry (MS)/MS analysis, data preprocessing and filtering, and a multivariate statistical analysis were performed on the samples. The results of the untargeted lipidomic analysis are provided in the available online: https://cdn.amegroups.cn/static/public/atm-21-3777-01.xlsx.
Transcriptomic analysis
There were 9 samples per group, which were mixed into 3 samples for the transcriptomic analysis. The samples were analyzed as described previously (20-24). Detailed steps are outlined in the Appendix 2. In short, after ribonucleic acid (RNA) extraction and qualification, the samples were subject to a transcriptomic analysis, library preparation for transcriptome sequencing, clustering and sequencing. The results of the transcriptomic analysis were verified by a quantitative polymerase chain reaction (qPCR) using sequence-specific primers. The results of the transcriptomic analysis and qPCR are provided in the available online: https://cdn.amegroups.cn/static/public/Appendix4-5.zip.
Statistics
Values are expressed as means ± standard error of the mean (SEM). Significant differences were assessed by t-tests. GraphPad Prism software (ver. 8.0; RRID:SCR_002798 GraphPad, La Jolla, CA, USA) was used to perform all the statistical analyses. A value of P<0.05 was considered significant.
Results
The histopathological effects of D-4F on BLM-induced pulmonary fibrosis
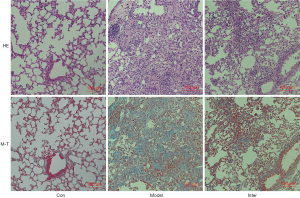
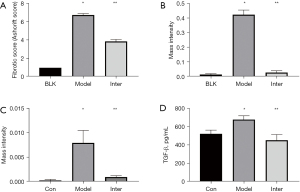
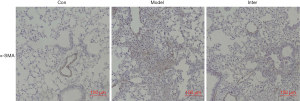
After instilling BLM into the mouse lungs, H&E and M-T staining of the Model Group showed significant pulmonary fibrosis. The lung structure showed severe distortion and collagen fibers were accumulated in the lung (see Figure 1). The Ashcroft score and image analysis of M-T staining in the Model group were significantly more severe than those of BLK group . However, the results of H&E and M-T staining revealed that the Inter Group showed significantly attenuated BLM-induced pulmonary fibrosis. The structure of the alveolar wall and diffuse fibrosis were reduced in the Inter Group. The Ashcroft score and the imaging analysis results of the M-T staining of the Inter Group were significantly lower than those of the Model Group (see Figure 2). The α-SMA is a kind of mesenchymal marker. α-SMA expression in the interstitial fibrosis was higher in Model Group than the Inter Group (see Figure 3), but was significantly more decreased in the Inter Group than the Model Group (see Figure 2). To gain insights into the pathological changes, the level of TGF-β1 in the serum was analyzed and summarized (see Figure 2). The results showed that the serum level of TGF-β1 in the Model Group was significantly higher than that in the BLK Group. The TGF-β1 level of the Inter Group was significantly lower compared to that of the Model Group .
The ayUntargeted lipidomic analysis in the D-4F group
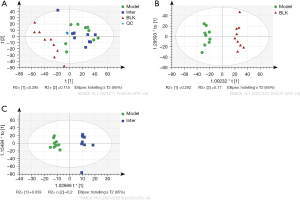
The lipidomic of every group was analyzed by a principal component analysis (PCA) and orthogonal partial least-squares discriminant analysis (OPLS-DA) plots (see Figure 4). An obvious separation was observed in the OPLS-DA score scatter plots between the BLK and Model Groups (R2Y: 0.977, Q2: 0.792, Q2 intercept: −0.376), and the Model and Inter Groups (R2Y: 0.957, Q2: 0.405, Q2 intercept: −0.291).
Using a variable importance in projection (VIP) score >1 and a P<0.05, we identified 1,100 differentially altered lipids (DALs) in the BLK and Model Groups and 92 DALs in the Model and Inter Groups. There were 35 common DALs in the 1,100 DALs and 92 DALs (see Figure 5). The 92 DALs of the Model and Inter Groups comprised 65 glycerophospholipids, 6 sphingolipids, 16 glycerides, and 5 other lipids. The glycerophospholipids contained the following 13 classes: phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidylglycerol (PG), phosphatidylethanol (PEt), phosphatidylethanolamine (PE), phosphatidylcholine (PC), lysophosphatidylserine (LPS), lysophosphatidylmethanol (LPMe), lysophosphatidylinositol (LPI), lysophosphatidylethanolamine (LPE), lysophosphatidylcholine (LPC), dimethylphosphatidylethanolamine (dMePE), and lysodimethylphosphatidylethanolamine (LdMePE). The sphingolipids contained the following 3 classes: phytosphingosine (phSM), sphingomyelin (SM), and Gangliosides (GM3). The glycerides contained the following 2 classes: diglyceride (DG) and triglyceride (TG). The other lipids contained fatty acid (FA), Digalactosyldiacylglycerol (DGDG), and Sulfoquinovosyldiacylglycerol (SQDG) (see Figure 6).
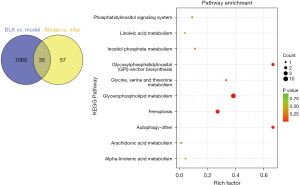
To characterize any related lung lipids altered by D-4F treatments, we conducted a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis to identify the top 10 significant pathways in the 35 common DALs (see Figure 5). The potential target “glycerophospholipid metabolism” pathway was the most highly enriched (10 converted lipids; P=7.662×10−11).
The transcriptomic analysis in the D-4F group
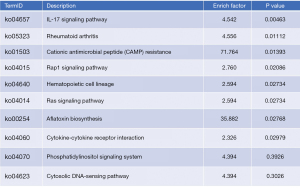
To determine the lipidomic alterations between the Model Group and the Inter Group, transcriptomic changes were assessed using RNA-sequencing. Using a fold change ≥2 or <2 and a P<0.05, we identified 2,179 DEGs in the BLK and Model Groups, and 217 DEGs in the Model and Inter Groups (see Figure 7). The top 10 significant enriched KEGG analysis pathways were shown (see Figure 8). The Ras signaling pathway (P=0.02734) was in these pathways, and the genes involved in this pathway were Fgf23, Fgfr3, Flt1, phospholipase A2 group 4c (Pla2g4c), Plce1, and Rapgef5.
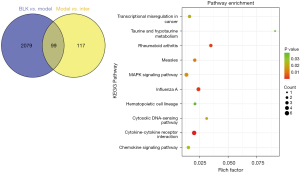
There were 99 common DEGs in the BLK, Model Group and Inter Groups. To identify the key pathways associated with D-4F, a KEGG analysis was performed on the 99 common DEGs. The top 10 significant pathways in these DEGs are shown in Figure 9. We chose the mitogen-activated protein kinase (MAPK) signaling pathway as a key pathway. This pathway was obviously enriched (4 gene counts; P=0.0152).
The lipid-transcript correlative analysis
Based on the results of the lipidomic and transcriptomic analysis of the Model and Inter Groups, the protein-protein interaction network (PPI network) was employed. This network was constructed from 217 DEGs and 92 DALs. The PPI network comprised 10 pathways, and we selected glycerophospholipid metabolism based on the results of the lipidomic analysis (see Figure 10).
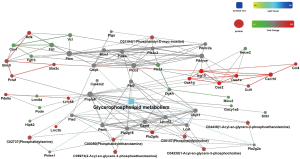
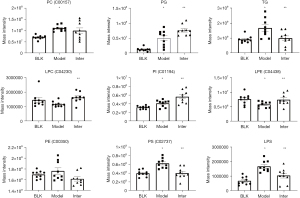
To conform to the PPI network of glycerophospholipid metabolism, the 35 common DALs in the D-4F group were divided into the following 9 classes: C00157, C04230, C00350, C01194, C02737, C04438, PG, LPS, and TG. Compared to that of the Model Group, the lipid classes of the Inter Group were significantly decreased in terms of TG (P=0.0189), C02737 (P=0.0013), and LPS (P=0.0081). and increased in terms of C04230 (P=0.0046), PG (P=0.0247), C01194 (P=0.0142), and C04438 (P=0.0423). C00157 and C00350 were no significant difference between the Model Group and the Inter Group (see Figure 11).
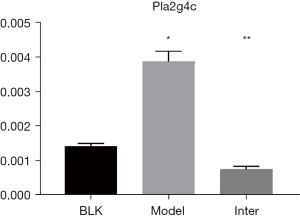
Pla2g4c is a crucial gene in the MAPK pathway, and glycerophospholipid metabolism. Pla2g4c is a calcium+-independent PLA2 family (25,26) and was significantly decreased in the Inter Group. Pla2g4c is a member of the PLA2 family, and pla2g2a, which is another member of the PLA2 family, has been reported to be increased in BLM-induced rat lungs (27). The results of the transcriptomic analysis revealed that pla2g4c was increased in the Model Group and decreased in the Inter Group. The changes of pla2g4c in the BLK, Model, and Inter Groups were verified by qPCR (see Figure 12). Overall, these data suggest that pla2g4c expression is related to IPF and could be a potential target of D-4F.
Discussion
IPF is a chronic fibrotic pulmonary disease that is accompanied by lipid alterations in the lung. Kim et al. (5) reported ApoA-1 was significantly decreased in the bronchoalveolar lavage fluid of IPF patients, and that treatment with ApoA-1 reduced inflammatory cells and collagen deposition. D-4F is an ApoA-1 mimetic that mimics the functions of ApoA-1. The results of our lipidomic and transcriptomics analysis showed that D-4F ameliorated the BLM-induced model of IPF via the adenosine monophosphate-activated protein kinase (AMPK) pathway and glycerophospholipid metabolism.
D-4F was designed to heal the disorder resulting from an ApoA-1 deficiency. Previous research has focused on the effects of D-4F on cardiovascular disorders. D-4F has been shown to improve the anti-inflammatory functions of HDLs (28) decrease atherosclerotic lesions (29), convert HDLs from pro-inflammatory to anti-inflammatory, and oxidize phospholipids (10,30). Bloedon et al. (31) was the first to report that an oral D-4F in humans was effective, was absorbed rapidly, and ameliorated the HDL inflammatory index in high-risk cardiovascular patients. Recently the effects of D-4F on non-cardiovascular disorders were examined and good results were achieved. Specifically, D-4F was found to decrease oligodendrocyte-progenitor cell death after stroke (32), inhibit the proliferation and tumorigenicity of epithelial ovarian cancer cells (33), ameliorate contrast media-induced oxidative injuries via the AMPK/protein kinase C (PKC) pathway (34), reduce inflammatory responses resulting from influenza infections (35). In experimental asthma D-4F decreased the expression of biomarkers of oxidative stress, attenuated recruitment of inflammatory cells and prevented collagen deposition (36). These functions result in the reduction of pulmonary stiffness and the decrease of airway hyperresponsiveness. EMT and macrophage alternative activation are involved in pulmonary fibrosis. In the TGF-β1-induced-AECs, D-4F increased the expression of E-cadherin and decreased the expression of vimentin (12). These suggested that D-4F inhibits TGF-β1 induced EMT. Alveolar M2 macrophage increased in the lung and bronchoalveolar lavage fluid from patients with IPF, which indicated that M2 macrophage had a key role in controlling fibrogenesis (37). D-4F suppressed IL-4 induced macrophage alternative activation, and reduced TGF-β1transcription and translation (38). D-4F inhibits EMT and macrophage alternative activation in vitro, indicating that D-4F may attenuate IPF in vivo.
An untargeted lipidomic analysis was conducted to examine the effects of D-4F on BLM-induced mice with a lipid metabolites disorder. Overall, we found that the lipids in the lung were significantly changed in BLM-induced and D-4F-treated lungs. In this study, there were 35 common DALs in the D-4F group. A KEGG pathway analysis of those DALs revealed a significant alteration in glycerophospholipid metabolism. The PG and PI levels of the DALs were increased in the Model and Inter Groups . PG is a major component of lung, and has been reported to interact with macrophages Monocyte differentiation antigen CD14 (CD14) and myeloid differentiation protein-2 (MD2) in vitro (39). Further, it was reported that PI binds to the CD14 plasma membrane and prevents monocyte activation by bacterial lipopolysaccharides (40). Saito et al. (15) conjectured that high levels of PG and PI counteract inflammation and thus ameliorate BLM-induced lung injury . Our study showed that PG and PI were more increased in the Inter Group than the Model Group, and D-4F can decrease inflammation in experimental asthma and influenza infection. Thus, D-4F increased PG and PI by reducing inflammation to improve BLM-induced lung injury.
Additionally, PS was significantly increased in the Model Group but decreased in the Inter Group. PS exists in all human cells, is an essential component on the inner leaflet of the cell membrane, and is externalized upon the induction of apoptosis, resulting in efferocytosis (41). The overexpression of PS enhances efferocytosis, leading to autoimmunity (42). Research has shown that apoptotic levels were elevated in the bronchoalveolar lavage macrophages of patients with IPF (43). Additionally, previous research has shown that the growing expression of TGF-β results from the uptake of apoptotic cells by macrophages, which is a growth factor in both anti-inflammatory and pro-fibrotic activities (44). Thus, in relation to the decreased levels of PS in the Inter Group, it may be that D-4F counteracted the apoptosis and suppressed macrophage activation.
The results of the transcriptomic and KEGG analyses showed that the MAPK pathway was involved in the ApoA-1-/- group and the D-4F group. MAPK is a cellular bioenergetic sensor metabolic regulator and plays an important role in growth differentiation and stress responses (45). The activated MAPK pathway is directly correlated with the severity of pulmonary fibrosis. In IPF patients, the MAPK pathway is activated in epithelial cells, smooth muscle cells, and fibroblasts (46). The TGF-β activated MAPK pathway is essential in the aberrant proliferation of pulmonary interstitial fibroblasts (47). Conversely, the specific inhibitor FR-167653, which inhibits MAPK signaling, ameliorated BLM-induced IPF in mice (48). D-4F decreased the level of TGF-β in the Model Group and inhibited the MAPK pathway to ameliorate BLM-induced lung injury. Additionally, the Ras signaling pathway was enriched in the Model and Inter Groups. The Ras signaling pathway plays an important role in regulating blood pressure and electrolyte balance. Recently, research has begun to unravel the role of Ras in inflammatory responses in the lungs (49). TGF-β is the most potent pro-fibrotic cytokine and acts downstream of Ang II in vascular smooth muscles, myofibroblasts, and macrophages (50). EMT, which is a hallmark of IPF, was induced by TGF-β via the RAS and MAPK pathway (51). Additionally, Renin inhibition by aliskiren has been reported to attenuate lung fibrosis, which decreases TGF-β and prevents myofibroblasts activation and differentiation in the BLM-induced IPF model (52). Overall, D-4F ameliorated BLM-induced lung injury and decreased the levels of TGF-β via the Ras/MAPK pathway.
Pla2g4c [also referred to as cytosolic PLA2gamma (cPLA2γ)] was identified as an ortholog cPLA2a belonging to the cPAL2 family and contains a lipase consensus sequence that lacks the C2 domain (53,54). cPLA2γ is a major enzyme involved in phospholipid AA remodeling (55). Additionally, cPLA2γ catalyzes CoA-dependent transacylation and exhibits lysophospholipase/transacylation activity, but it prefers lysophospholipase/transacylation to CoA-dependent transacylation reaction as acyl donors (53,54,56). Yamashita et al. suggested that purified cPLA2γ catalyzes acyltransferase activity between 2 LPC molecules to form PC and glycerophosphocholine, and between 2 molecules of LPE to form PE and glycerophosphoethanolamine (53). In this study, the level of pla2g4c decreased, while the levels of LPC and LPE increased, which suggests that pla2g4c plays a crucial role in the treatment of D-4F on BLM-induced IPF.
In conclusion, this study demonstrated that D-4F ameliorating BLM-induced IPF is associated with glycerophospholipid metabolism and the Ras/MAPK pathway. Further, pla2g4c may be the core genes that alleviate BLM-induced IPF after D-4F treatment. Our results identified potential targets for treating IPF.
Acknowledgments
Funding: This work was supported by the Research Award Fund for Outstanding Young Scientists of Shandong Province (BS2015YY043), the Natural Science Foundation of the Shandong Province (ID-ZR2017MH122), and the Shandong Medicine and Health Science and Technology Development Plan Project (2014WS0139).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3777
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-3777
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3777). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (No.: 21109) granted by Laboratory Animal Ethical and Welfare Committee of Shandong University Cheeloo College of Medicine, in compliance with Shandong University Cheeloo College of Medicine guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Datta A, Scotton CJ, Chambers RC. Novel therapeutic approaches for pulmonary fibrosis. Br J Pharmacol 2011;163:141-72. [Crossref] [PubMed]
- King TE Jr, Pardo A, Selman M. Idiopathic pulmonary fibrosis. Lancet 2011;378:1949-61. [Crossref] [PubMed]
- Tham T. MLKL-dependent epithelial-to-mesenchymal transition in nasopharyngeal carcinoma: a novel finding and avenues for future research. Ann Transl Med 2020;8:153. [Crossref] [PubMed]
- Willis BC, Liebler JM, Luby-Phelps K, et al. Induction of epithelial-mesenchymal transition in alveolar epithelial cells by transforming growth factor-beta1: potential role in idiopathic pulmonary fibrosis. Am J Pathol 2005;166:1321-32. [Crossref] [PubMed]
- Kim TH, Lee YH, Kim KH, et al. Role of lung apolipoprotein A-I in idiopathic pulmonary fibrosis: antiinflammatory and antifibrotic effect on experimental lung injury and fibrosis. Am J Respir Crit Care Med 2010;182:633-42. [Crossref] [PubMed]
- Cuchel M, Rader DJ. Macrophage reverse cholesterol transport: key to the regression of atherosclerosis? Circulation 2006;113:2548-55. [Crossref] [PubMed]
- Yao X, Gordon EM, Figueroa DM, et al. Emerging Roles of Apolipoprotein E and Apolipoprotein A-I in the Pathogenesis and Treatment of Lung Disease. Am J Respir Cell Mol Biol 2016;55:159-69. [Crossref] [PubMed]
- Lee Eh, Lee EJ. Overexpression of apolipoprotein A1 in the lung abrogates fibrosis in experimental silicosis. PLoS One 2013;8:e55827 [Crossref] [PubMed]
- Navab M, Anantharamaiah GM, Reddy ST, et al. Human apolipoprotein AI mimetic peptides for the treatment of atherosclerosis. Curr Opin Investig Drugs 2003;4:1100-4. [PubMed]
- Navab M, Anantharamaiah GM, Reddy ST, et al. Apolipoprotein A-I mimetic peptides. Arterioscler Thromb Vasc Biol 2005;25:1325-31. [Crossref] [PubMed]
- Navab M, Anantharamaiah GM, Reddy ST, et al. Peptide Mimetics of Apolipoproteins Improve HDL Function. J Clin Lipidol 2007;1:142-7. [Crossref] [PubMed]
- You J, Wang J, Xie L, et al. D-4F, an apolipoprotein A-I mimetic, inhibits TGF-β1 induced epithelial-mesenchymal transition in human alveolar epithelial cell. Exp Toxicol Pathol 2016;68:533-41. [Crossref] [PubMed]
- Percie du Sert N, Ahluwalia A, Alam S, et al. Reporting animal research: Explanation and elaboration for the ARRIVE guidelines 2.0. PLoS Biol 2020;18:e3000411 [Crossref] [PubMed]
- Percie du Sert N, Hurst V, Ahluwalia A, et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. J Cereb Blood Flow Metab 2020;40:1769-77. [Crossref] [PubMed]
- Saito K, Tanaka N, Ikari J, et al. Comprehensive lipid profiling of bleomycin-induced lung injury. J Appl Toxicol 2019;39:658-71. [Crossref] [PubMed]
- Ashcroft T, Simpson JM, Timbrell V. Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J Clin Pathol 1988;41:467-70. [Crossref] [PubMed]
- Chen S, Hoene M, Li J, et al. Simultaneous extraction of metabolome and lipidome with methyl tert-butyl ether from a single small tissue sample for ultra-high performance liquid chromatography/mass spectrometry. J Chromatogr A 2013;1298:9-16. [Crossref] [PubMed]
- Tang H, Wang X, Xu L, et al. Establishment of local searching methods for orbitrap-based high throughput metabolomics analysis. Talanta 2016;156-157:163-71. [Crossref] [PubMed]
- Xu L, Wang X, Jiao Y, et al. Assessment of potential false positives via orbitrap-based untargeted lipidomics from rat tissues. Talanta 2018;178:287-93. [Crossref] [PubMed]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal 2011;17:10-2. [Crossref]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357-60. [Crossref] [PubMed]
- Pertea M, Pertea GM, Antonescu CM, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 2015;33:290-5. [Crossref] [PubMed]
- Frazee AC, Pertea G, Jaffe AE, et al. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol 2015;33:243-6. [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Underwood KW, Song C, Kriz RW, et al. A novel calcium-independent phospholipase A2, cPLA2-gamma, that is prenylated and contains homology to cPLA2. J Biol Chem 1998;273:21926-32. [Crossref] [PubMed]
- Lucas KK, Dennis EA. The ABC's of Group IV cytosolic phospholipase A2. Biochim Biophys Acta 2004;1636:213-8. [Crossref] [PubMed]
- Bauer Y, Tedrow J, de Bernard S, et al. A novel genomic signature with translational significance for human idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol 2015;52:217-31. [Crossref] [PubMed]
- Navab M, Anantharamaiah GM, Reddy ST, et al. Apolipoprotein A-I mimetic peptides and their role in atherosclerosis prevention. Nat Clin Pract Cardiovasc Med 2006;3:540-7. [Crossref] [PubMed]
- Getz GS, Reardon CA. Apolipoprotein A-I and A-I mimetic peptides: a role in atherosclerosis. J Inflamm Res 2011;4:83-92. [Crossref] [PubMed]
- Anantharamaiah GM, Mishra VK, Garber DW, et al. Structural requirements for antioxidative and anti-inflammatory properties of apolipoprotein A-I mimetic peptides. J Lipid Res 2007;48:1915-23. [Crossref] [PubMed]
- Bloedon LT, Dunbar R, Duffy D, et al. Safety, pharmacokinetics, and pharmacodynamics of oral apoA-I mimetic peptide D-4F in high-risk cardiovascular patients. J Lipid Res 2008;49:1344-52. [Crossref] [PubMed]
- Cui X, Chopp M, Zacharek A, et al. D-4F Decreases White Matter Damage After Stroke in Mice. Stroke 2016;47:214-20. [Crossref] [PubMed]
- Ganapathy E, Su F, Meriwether D, et al. D-4F, an apoA-I mimetic peptide, inhibits proliferation and tumorigenicity of epithelial ovarian cancer cells by upregulating the antioxidant enzyme MnSOD. Int J Cancer 2012;130:1071-81. [Crossref] [PubMed]
- Guo Y, Li W, Qian M, et al. D-4F Ameliorates Contrast Media-Induced Oxidative Injuries in Endothelial Cells via the AMPK/PKC Pathway. Front Pharmacol 2021;11:556074 [Crossref] [PubMed]
- Van Lenten BJ, Wagner AC, Navab M, et al. D-4F, an apolipoprotein A-I mimetic peptide, inhibits the inflammatory response induced by influenza A infection of human type II pneumocytes. Circulation 2004;110:3252-8. [Crossref] [PubMed]
- Nandedkar SD, Weihrauch D, Xu H, et al. D-4F, an apoA-1 mimetic, decreases airway hyperresponsiveness, inflammation, and oxidative stress in a murine model of asthma. J Lipid Res 2011;52:499-508. [Crossref] [PubMed]
- Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med 2012;18:1028-40. [Crossref] [PubMed]
- Song X, Shi Y, You J, et al. D-4F, an apolipoprotein A-I mimetic, suppresses IL-4 induced macrophage alternative activation and pro-fibrotic TGF-β1 expression. Pharm Biol 2019;57:470-6. [Crossref] [PubMed]
- Kuronuma K, Mitsuzawa H, Takeda K, et al. Anionic pulmonary surfactant phospholipids inhibit inflammatory responses from alveolar macrophages and U937 cells by binding the lipopolysaccharide-interacting proteins CD14 and MD-2. J Biol Chem 2009;284:25488-500. [Crossref] [PubMed]
- Wang PY, Kitchens RL, Munford RS. Phosphatidylinositides bind to plasma membrane CD14 and can prevent monocyte activation by bacterial lipopolysaccharide. J Biol Chem 1998;273:24309-13. [Crossref] [PubMed]
- Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys 2010;39:407-27. [Crossref] [PubMed]
- Asano K, Miwa M, Miwa K, et al. Masking of phosphatidylserine inhibits apoptotic cell engulfment and induces autoantibody production in mice. J Exp Med 2004;200:459-67. [Crossref] [PubMed]
- Morimoto K, Janssen WJ, Terada M. Defective efferocytosis by alveolar macrophages in IPF patients. Respir Med 2012;106:1800-3. [Crossref] [PubMed]
- Fadok VA, Bratton DL, Konowal A, et al. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J Clin Invest 1998;101:890-8. [Crossref] [PubMed]
- Cobb MH, Goldsmith EJ. How MAP kinases are regulated. J Biol Chem 1995;270:14843-6. [Crossref] [PubMed]
- Yoshida K, Kuwano K, Hagimoto N, et al. MAP kinase activation and apoptosis in lung tissues from patients with idiopathic pulmonary fibrosis. J Pathol 2002;198:388-96. [Crossref] [PubMed]
- Khalil N, Xu YD, O'Connor R, et al. Proliferation of pulmonary interstitial fibroblasts is mediated by transforming growth factor-beta1-induced release of extracellular fibroblast growth factor-2 and phosphorylation of p38 MAPK and JNK. J Biol Chem 2005;280:43000-9. [Crossref] [PubMed]
- Matsuoka H, Arai T, Mori M, et al. A p38 MAPK inhibitor, FR-167653, ameliorates murine bleomycin-induced pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2002;283:L103-12. [Crossref] [PubMed]
- Jia H. Pulmonary Angiotensin-Converting Enzyme 2 (ACE2) and Inflammatory Lung Disease. Shock 2016;46:239-48. [Crossref] [PubMed]
- Uhal BD, Kim JK, Li X, et al. Angiotensin-TGF-beta 1 crosstalk in human idiopathic pulmonary fibrosis: autocrine mechanisms in myofibroblasts and macrophages. Curr Pharm Des 2007;13:1247-56. [Crossref] [PubMed]
- Su J, Morgani SM, David CJ, et al. TGF-β orchestrates fibrogenic and developmental EMTs via the RAS effector RREB1. Nature 2020;577:566-71. [Crossref] [PubMed]
- Asker SA, Mazroa SA, Boshra V, et al. Biochemical and histological impact of direct renin inhibition by aliskiren on myofibroblasts activation and differentiation in bleomycin induced pulmonary fibrosis in adult mice. Tissue Cell 2015;47:373-81. [Crossref] [PubMed]
- Yamashita A, Tanaka K, Kamata R, et al. Subcellular localization and lysophospholipase/transacylation activities of human group IVC phospholipase A2 (cPLA2gamma). Biochim Biophys Acta 2009;1791:1011-22. [Crossref] [PubMed]
- Yamashita A, Hayashi Y, Matsumoto N, et al. Coenzyme-A-Independent Transacylation System; Possible Involvement of Phospholipase A2 in Transacylation. Biology (Basel) 2017;6:23. [Crossref] [PubMed]
- Lebrero P, Astudillo AM, Rubio JM, et al. Cellular Plasmalogen Content Does Not Influence Arachidonic Acid Levels or Distribution in Macrophages: A Role for Cytosolic Phospholipase A2γ in Phospholipid Remodeling. Cells 2019;8:799. [Crossref] [PubMed]
- Yamashita A, Kamata R, Kawagishi N, et al. Roles of C-terminal processing, and involvement in transacylation reaction of human group IVC phospholipase A2 (cPLA2gamma). J Biochem 2005;137:557-67. [Crossref] [PubMed]




