Functional assessment of a novel COL4A5 splicing site variant in a Chinese X-linked Alport syndrome family
Introduction
Alport syndrome (AS) is an inherited renal disorder characterized by hematuria, proteinuria, and progressive renal failure, and occasionally accompanied by ocular abnormalities and hearing loss (1). According to previous reports, 85% of the patients inherit COL4A5 mutations in an X-linked dominant manner, whereas autosomal recessive AS caused by mutations in COL4A3 or COL4A4 accounts for 15% of the cases (2).
The COL4A5 gene is located on chromosome Xq22 and encodes the α5 chain of type IV collagen, the primary constituent of the GBM, cochlea, and eye. Variants in the COL4A5 gene prevent normal triple-helix formation and secretion of type IV collagen, resulting in defects in GBM (3). According to recent statistics, at least 1,900 COL4A5 variants have been reported (4). In XLAS, about 40% of all mutations are missense, 10% are splicing mutations, 7% are nonsense mutations, and a further 30% result in a frame-shift and downstream nonsense change (5). Although the genotype-phenotype correlations in XLAS have not been completely illuminated, it is generally believed that missense mutations exhibit milder phenotypes than truncating mutations (6-8).
Regarding splice site mutations, research suggests that these cases tend to show moderate severity (6,8,9). In this study, we used NGS to identify a novel splicing site variant downstream of COL4A5 intron 18 in an XLAS family with both male and female members affected. We then further analyzed the consequences of this gene variant on the translated protein using in silico and in vitro approaches.
We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3523).
Methods
Ethical statement
The study protocol was conducted based on the principles of the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee on Biomedical Research, West China Hospital of Sichuan University (No. 2021-1005). Since this study is a retrospective research without any intervention, the informed consent was waived.
Clinical data
Referring physicians provided deidentified documentation regarding the clinical histories and laboratory data of the family members. A renal biopsy of the proband was performed at West China Hospital of Sichuan University; the proband’s sister underwent renal biopsy at her local hospital, and we collected the pathological reports of her biopsy.
Whole-exome sequencing (WES) and variation analysis
Sample collection
The patients provided informed consent and ethylenediaminetetraacetate (EDTA)-treated peripheral blood was collected.
DNA extraction
According to the manufactural instructions, the genomic DNA was extracted using the Blood Genome Column Medium Extraction Kit (Kang Wei Shi Ji, China). The extracted DNA samples were subject to quality control using Qubit 2.0 fluorimeter and electrophoresis with 0.8% agarose gel before progressing further with the protocol.
Whole exome library construction
Protein-coding exome enrichment was performed using xGen Exome Research Panel v1.0 (IDT, Iowa, USA). This panel consists of 429,826 individually synthesized and quality-controlled probes that target 39 Mb protein-coding regions (19,396 genes) of the human genome and cover 51 Mb of end-to-end tiled probe space.
Sequencing
High-throughput sequencing was performed by Illumina NovaSeq 6000 series sequencer (PE150), and 99% or more of the target sequences were sequenced. The sequencing process was performed by Beijing Chigene Translational Medicine Research Center Co., Ltd., 100875, Beijing.
Bioinformatics analysis
Quality control
Raw data were processed by Fastp toolkit software to perform quality control, including adapters removing and low-quality reads filtering.
Variants calling
The paired-end reads were performed using Burrows-Wheeler Aligner (BWA) to the Ensemble GRCh37/hg19 reference genome. Base quality score recalibration, together with single nucleotide polymorphism (SNP) and short indel calling, was conducted using the Genome Analysis Toolkit. SNPs and indels were screened to obtain high-quality and reliable variants according to the sequencing depth and variant quality.
Annotation and pathogenicity prediction of variants
The online system independently developed by Chigene (www.chigene.org) was used with the database to annotate minor allele frequencies (MAFs) and American College of Medical Genetics and Genomics (ACMG) practice guideline-based pathogenicity of every yielded gene variant. The system also provides a serial software package for conservative analysis and protein product structure prediction. The databases for MAFs annotation include 1,000 genomes, the Single Nucleotide Polymorphism Database (dbSNP), Exome Sequencing Project (ESP), the Exome Aggregation Consortium (ExAC), and the Chigene in-house MAFs database; Provean, Sift, Polypen2_hdiv, Polypen2_hvar, Mutationtaster, M-Cap, and Revel software packages were used to predict variation in the protein product structure. In line with the ACMG guideline, OMIM, HGMD, and ClinVar databases were used to validate the pathogenicity of every variant. To predict a functional change of variants on the splicing sites, MaxEntScan, dbscSNV, and GTAG software packages were used instead of product structure prediction software.
Mutation confirmation by sanger sequencing
Sanger sequencing was used to confirm the mutation in COL4A5 of the proband and the other family members. The primers for amplification were: forward 5'-TAGTTTTATATGCTTCGGGCTCA-3' and reverse 5'-GACACTATTCACTTGGGCTTT-3'. The PCR product was sequenced by ABI 3730XL and aligned with reference mRNA transcript (NM_033380) by DNASTAR software.
Splicing analysis
In order to investigate the possible consequences of the mutation, we submitted the DNA sequence from Exon 18 to Exon 19 of COL4A5 gene (about 2.3 kb) to Alternative Splice Site Predictor (ASSP, http://wangcomputing.com/assp/). To further verify the abnormal splice site, we amplified the COL4A5 gene fragment from Exon 18 to Exon 19 by using human genome DNA as a template. The polymerase chain reaction (PCR) and primers, and under which conditions, were listed in Table 1 (PCR-1). PCR product was inserted into a T-clone vector pEASY-T1 (Transgene, Beijing, China). The construct was used to transform E. coli TOP10 strain, and an ampicillin plate screening was performed. Plasmids extracted from E. coli colonies were pre-identified by 0.8% agarose gel electrophoresis after HindIII-EcoRI double enzyme digestion. The inserted fragment in the positive plasmid was further identified by DNA sequencing. The plasmid with the correct sequence was named as pEASY_COL4A5_Exon 18–19. In order to obtain the mutation fragment, a long downstream primer (primer 3, see Table 1) was designed to induce a point-mutation. The pEASY_COL4A5_Exon 18–19 was used as a template; the primer pair was primer 1 and primer 3 (Table 1), and the PCR condition was listed in Table 1 (PCR-2). Using the same protocol, the mutated fragment was cloned into the pEASY-T1 vector and identified by DNA sequencing. The plasmid with the expected mutative fragment was named pEASY_COL4A5_Exon 18–19m. The COL4A5_Exon 18–19 fragment or Exon 18–19m fragment was then transferred into a eukaryotic expression vector, pcDNA-3.1(+), by using Hind III and Not I endonuclease sites to form plasmid pcDNA3.1_COL4A5_Exon 18–19 or plasmid pcDNA3.1_COL4A5_Exon 18–19m. Plasmid pcDNA3.1_COL4A5_Exon 18-19 or plasmid pcDNA3.1_COL4A5_Exon 18–19m was transfected into 293T cells by using Lipofectamine 3000TM for transcription. At 24 h post transfection, all the RNA was separately extracted from the cells using a Simply P Total RNA Extraction Kit (BioFlux, China). This was followed by being reverse transcribed into cDNA with a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc, US) using a random hexamer primer. PCR was carried out to amply the cDNAs transcribed from plasmid pcDNA3.1_COL4A5_Exon 18–19 or plasmid pcDNA3.1_COL4A5_Exon 18-19m with primer 1 and primer 2 (PCR-3 in Table 1). PCR products were assayed by 1.2% agarose gel electrophoresis. Bands about 300 bp were purified by a Gel Extraction Kit and then sequenced.
Table 1
| PCR | Primer pair | PCR condition | Product length |
|---|---|---|---|
| PCR-1 | Primer 1 & Primer 2 | Melting temperature: 58 °C; extending time: 120 s; cycling times: 30 | 2.4 kb |
| PCR-2 | Primer 1 & Primer 3 | Melting temperature: 58 °C; extending time: 120 s; cycling times: 30 | 2.4 kb |
| PCR-3 | Primer 1 & Primer 2 | Melting temperature: 58 °C; extending time: 20 s; cycling times: 30 | 310 bp |
Primer 1, CTGAGACACCATCACAGGTTAGGCTTA; Primer 2, CTCGCTCTCCTTTTTCTCCAGGCA; Primer 3, CTCGCTCTCCTTTTTCTCCAGGCAACCCAGGCAACCCAATGTTTCCTTTTTCTCCTATAGTTATACCAGTCCCAGGTCTAGGAATTACCCTTATTACCAAAGAAAAAAAAAATAAAGAAATGCAAAGAAGA.
Immunohistochemical stain for collagen IV α5 in GBM
The proband kidney biopsy collected was formalin-fixed and embedded in paraffin using routine procedures. Immunohistochemical staining was performed on 2 µm paraffin sections by using rabbit anti-human collagen IV α5 chains antibody (ab231957, Abcam, USA). The control used was tissue from another patient who was diagnosed with minimal change nephropathy.
Results
Family clinical findings
The pedigree of this family is shown as Figure 1A. Individual II-1 (22 years old) is the proband, who presented microhematuria and proteinuria at the age of 18. He was admitted into West China Hospital and diagnosed with suspected AS according to the lamellated appearance of the GBM following renal biopsy. To confirm the diagnosis of AS, all the close family members of the proband were advised to undergo an investigation for renal disease. The proband’s sister (Individual II-2, 24 years old) also presented microhematuria, but proteinuria was absent. Then she underwent renal biopsy in a local hospital. Electron microscopy demonstrated thinning of GBM thickness (126–240 nm) with areas of podocyte foot processes fusion. The mother of the proband (Individual I-2, 46 years old) presented elevated serum creatine (1.7 mg/dL), along with both microhematuria and proteinuria. She had not been aware of her renal disorder, except for mild hypertension. Renal biopsy was not undertaken in the mother as ultrasonography confirmed kidney atrophy. The maternal grandfather passed away many years ago due to an unknown cause, and the maternal grandmother had preserved renal function and normal urinalysis results. The father of the proband (Individual I-1, 47 years old) had no sign of kidney disease. The affected members of this family are not known to have hearing loss or ocular abnormalities typical of Alport syndrome. In addition, this family denied any other consanguineous relatives affected by renal disease.

Genetic analysis
WES and variation analysis was performed on the proband and all his close family members. This revealed a novel splice variant c.1033-2A>G in intron 18 of COL4A5, located at the canonical splice site in the proband. Sanger sequencing confirmed that this variant is segregated with disease (Figure 1B). This variant was not found in the unaffected father of the proband. Both the mother and sister of the proband were heterozygous for this mutation. This mutation was not found previously in the public databases dbSNP, ExAC and 1,000 Genome databases; this was a rare variation. The software-predicted location of the nucleic acid conservative sequence showed that this site was in a conservative region. The splicing site harmfulness prediction software indicated that the site was deleterious. According to the variant interpretation guidelines of the ACMG (10), this variant was likely pathogenic.
Pathologic examination
Kidney pathology was examined in the proband. Light microscopy study found focal glomerulosclerosis and some segmental vacuolar degeneration of the GBM in the index patient. Partial tubular epithelial cells were swollen, and some fibrosis was found in the interstitium (Figure 2A,2B). Electron microscopic study revealed uneven thickening of GBM, with the splitting of the dense layer. Diffuse effacement of foot processes was seen (Figure 2C,2D).

In silico variant analysis
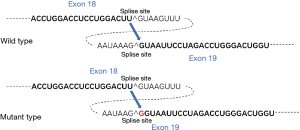
The mutated splice site between exon 18 and exon 19 was predicted by the Alternative Splice Site Predictor. It is understood that exon-intron splicing sites need to follow the rule that at the front end of the intron sequence is GU and at the back end is AG. In this variant case, the acceptor sequence at the front of exon 19 is changed from TAAAG^GTAA to TAAG^GGTAA, which results in an additional G insertion between exon 18 and exon 19 in the mature COL4A5 mRNA (Figure 3), subsequently leading to a frameshift and premature termination on translation.
In vitro splicing analysis
The complete experimental process for in vitro splicing analysis is shown in Figure 4A. In Figure 4B, a band of approximately 2.4 kb was amplified from human genome DNA with primer 1 and primer 2, which was then successfully inserted into the T-clone vector pEASY-T1. HindIII-EcoRI double enzyme digestion of pEASY_COL4A5_Exon 18–19 released a 2 kb band (Figure 4C), which indicated that cloned fragment was inserted in the right direction. Using pEASY_COL4A5_Exon 18–19 as a template, a mutated 2.4 kb fragment was created by primer 1 and primer 3 and then inserted into the pEASY-T1 vector to form the pEASY_COL4A5_Exon 18–19m plasmid. Inserted fragments were then transferred to the pcDNA3.1(+) vector to form plasmid pcDNA3.1_COL4A5_Exon 18–19 and plasmid pcDNA3.1_COL4A5_Exon 18–19m (Figure 4D). Target cDNA fragments (about 300 bp) were amplified by using primer 1 and primer 4 (Figure 4E), and then sequenced. As shown in Figure 4F, mRNA stitching product from plasmid pcDNA3.1_COL4A5_Exon 18–19 or plasmid pcDNA3.1_COL4A5_Exon 18–19m was assayed successfully, and a frameshift mutation (a G inserted) was clearly identified in the mRNA from pcDNA3.1_COL4A5_Exon 18–19m.

Collagen IV α5 stain in GBM
Unexpectedly, we observed positive collagen IV α5 immunohistochemical staining of GBM in the sections of both the control and the index patient (Figure 5). This result revealed that the α5 collagen IV chain was present in the GBM.

Discussion
This study reports a novel splice region variant in the COL4A5 gene in a family with both males and females affected by kidney disease. The proband and his sister seemed to inherit the disorder from their mother, and demonstrates a classic X-linked pattern of inheritance. In the past, it was thought that COL4A5 mutations resulted in abnormal type IV collagen-α5 chain expression, typically with a complete absence of α5(IV) in the GBM and Bowman’s capsule in men, and a mosaic expression pattern in women (11). Today, it has been demonstrated that a mild phenotype still exists in some males with XLAS due to missense or in-frame mutations (12). Splice site mutations are common in X-linked Alport syndrome. They can cause both truncating and nontruncating mutations, including in-frame deletions or in-frame mutations (13). Recently, it has been revealed that renal prognosis differs significantly for patients with truncating versus nontruncating splicing abnormalities. The median age for developing end-stage renal disease (ESRD) was 20 years among patients with truncating mutations and 29 years among patients with nontruncating mutations (14). Hence, it is important to clarify the diverse consequences of splice site mutations for evaluating the patient’s prognosis.
In this study, we conducted transcription analysis to identify a frameshift mutation caused by a novel splice region variant, c.1033-2(IVS 18) A>G, in COL4A5 intron 18, and verified this splicing result through molecular biology in a cell model. Theoretically, such a variant usually leads to creating a premature termination codon and results in a truncated protein product. However, we were limited by the number of kidney puncture samples, so we could not determine the resulting truncated protein directly from the biopsy of the index patient. We demonstrated the expression of α5(IV) in the GBM of the proband by immunohistology. Negative staining in the GBM was predicted because the antibody used only targets the AA1461-1685 on wild-type COL4A5. Unexpectedly, both the proband and the control images showed similar positive staining of α5(IV) in the GBM and Bowman’s capsule. Previously, it was reported that over 20% of male XLAS patients had positive α5(IV) staining. They exhibited non-truncating mutations or somatic mosaic mutations in COL4A5 and possessed milder clinical manifestations that involved lower urinary protein levels and later-onset ESRD (15). According to the RNA splicing analysis, this novel splice region variant results in a truncated protein product with only 347 amino acids caused by a frameshift. However, it is difficult to understand the positive immunohistology in tissue samples from the proband when using an antibody which only targets the C-terminal of wild-type COL4A5.
Further analysis on mutated COL4A5 mRNA showed there might be another potential open reading frame (ORF) beginning at 908 bp from the first start codon (ORF2 in Figure 6A), which could code a “head-missing” COL4A5 with 1,383 amino acids (see Figure 6B). A recent study showed alterations in translation initiation in eukaryotic mRNA might be a pervasive phenomenon and act as an important regulation (16). Another possibility is that some alternative splicing was performed by skipping intron 18 and intron 19 on this mutation gene, thus translating a protein that lacks an intermediate peptide fragment. Theoretically, because of the triple-helical region missing, the truncated protein product with only 347 amino acids would thoroughly lose the bio-function of collagen and lead to a serious symptom. However, the proband in this case exhibits a much milder symptom compared with other typical XLAS patients implicated by truncating mutation on the COL4A5 gene. Together with the positive immunohistology result of COL4A5 in proband tissue samples, we postulated that the protein products translated by these mutated mRNAs might still keep the partial biological functions and immunogenicity of COL4A5, which might explain the milder symptoms of the proband compared with other typical XLAS patients implicated by truncating mutation on the COL4A5 gene. These hypotheses need further verification by mRNA sequencing and western-blot assay in patient biopsies or a mutation cell model. Therefore, due to the nonspecific findings by α5 staining, we can only use it as an auxiliary diagnostic method for XLAS.

Nonspecific pathologic findings in Alport syndrome have been observed in many studies (17). We also noticed that the proband was first diagnosed as focal global and segmental glomerulosclerosis (FSGS) by light microscope examination. Whether there is common underlying pathogenesis is still unknown. The formation of collagen IV in the endoplasmic reticulum of podocytes is considered to be disturbed by the COL4 mutation. Nowadays, it is considered that absent or dysfunctional α3α4α5 (IV) collagen heterotrimer is probably associated with podocytes defective adherence and accelerated detachment from GBM, which contributes to the FSGS pattern of pathologic change in AS (18). Previous studies also reported that COL4 gene variants are associated with primary familial FSGS, but whether this pathological lesion is secondary to the GBM pathology remains controversial (19-21). Regardless, it is important to differentiate Alport syndrome from primary glomerulonephritis, especially in those with a family history. Further electron microscopic examination and genetic testing are necessary for suspected cases of the disease.
XLAS has previously been reported to show a clear genotype-phenotype linkage (22). Male patients are usually severely affected, whereas heterozygous female carriers possess a broad spectrum of symptoms from isolated hematuria to ESRD (23,24). However, this view has been challenged in recent years. Hashimura et al. reported that nearly 30% of male patients with XLAS in their study showed α5(IV) expression in kidney glomeruli and displayed milder phenotypes, including milder proteinuria, later onset of ESRD, and less occurrence of hearing loss (15). Many of these patients had nontruncating mutations resulting from splice site mutations. Even for truncating mutations, a mild phenotype is still a possibility (13). For AS in the Chinese population, although there are some literature reports, most of them are case studies. Observational studies that include a large sample size are still lacking. As shown by the available information, similar clinical phenotypes and genotypic characteristics to those reported in foreign patients were observed.
At the time of writing, the 24-year-old male proband in our study had achieved clinical remission (24-h urine total protein <500 mg and normal serum creatine) following therapy, including glucocorticoids, cyclosporine and irbesartan (renin-angiotensin-aldosterone system inhibitor). It seems that this patient had a relatively milder phenotype. In addition, the two female patients in this family had different clinical phenotypes. The mother had developed ESRD by the age of 45, while the daughter only has mild microscopic hematuria, which indicates a benign prognosis. Today, the underlying mechanisms for the different phenotypes in XLAS have not been completely clarified. Some researchers have found that certain mutations likely lead to the expression of both abnormal and normal transcription. The different frequency of normally spliced transcripts could explain the disparity in disease severity among the affected individuals in a family (25). Other studies consider the phenotypic variation among affected females is attributable to the extent of inactivation of the normal X chromosome (26), also called lyonization. Lyonization produces a mosaic distribution of the mutant collagen IV α5 chain in the female kidney and skin (26). Analysis of X inactivation may help understand phenotypic variations.
This study expands the mutational spectrum for the COL4A5 gene associated with XLAS and enriches our understanding of genotype-phenotype correlations in this disease. Due to its nonspecific clinical and pathologic manifestations, misdiagnosis of this disease is not uncommon. Patients with a family history of renal disorder should be encouraged to undergo genetic analysis to confirm the diagnosis of Alport syndrome. Furthermore, genetic counseling should be offered to clarify the probability of disease transmission and provide education about reproductive options.
Acknowledgments
Funding: This work was supported by the Science and Technology Department of Sichuan Province (no. 2021YFH0061), 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYGD18014).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3523
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-3523
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3523). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol was conducted based on the principles of the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee on Biomedical Research, West China Hospital of Sichuan University (No. 2021-1005). Since this study is a retrospective research without any intervention, the informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kruegel J, Rubel D, Gross O. Alport syndrome--insights from basic and clinical research. Nat Rev Nephrol 2013;9:170-8. [Crossref] [PubMed]
- Zhu F, Li W, Li Z, et al. Identification of a Novel COL4A4 Variant in Compound-Heterozygous State in a Patient With Alport Syndrome and Histological Findings Similar to Focal Segmental Glomerulosclerosis (FSGS). Front Genet 2019;9:748. [Crossref] [PubMed]
- Deltas C, Pierides A, Voskarides K. Molecular genetics of familial hematuric diseases. Nephrol Dial Transplant 2013;28:2946-60. [Crossref] [PubMed]
- Miner JH, Baigent C, Flinter F, et al. The 2014International Workshop on Alport Syndrome. Kidney Int 2014;86:679-84. [Crossref] [PubMed]
- Savige J, Ariani F, Mari F, et al. Expert consensus guidelines for the genetic diagnosis of Alport syndrome. Pediatr Nephrol 2019;34:1175-89. [Crossref] [PubMed]
- Bekheirnia MR, Reed B, Gregory MC, et al. Genotype-phenotype correlation in X-linked Alport syndrome. J Am Soc Nephrol 2010;21:876-83. [Crossref] [PubMed]
- Gross O, Netzer KO, Lambrecht R, et al. Meta-analysis of genotype-phenotype correlation in X-linked Alport syndrome: impact on clinical counselling. Nephrol Dial Transplant 2002;17:1218-27. [Crossref] [PubMed]
- Jais JP, Knebelmann B, Giatras I, et al. X-linked Alport syndrome: natural history in 195 families and genotype- phenotype correlations in males. J Am Soc Nephrol 2000;11:649-57. [Crossref] [PubMed]
- Chen C, Lu CX, Wang Q, et al. A Novel Splicing Mutation Identified in a Chinese Family with X-linked Alport Syndrome Using Targeted Next-Generation Sequencing. Genet Test Mol Biomarkers 2016;20:203-7. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Kashtan CE. Alport syndrome and thin glomerular basement membrane disease. J Am Soc Nephrol 1998;9:1736-50. [Crossref] [PubMed]
- Nozu K, Takaoka Y, Kai H, et al. Genetic background, recent advances in molecular biology, and development of novel therapy in Alport syndrome. Kidney Res Clin Pract. 2020;39:402-13. [Crossref] [PubMed]
- Nozu K, Vorechovsky I, Kaito H, et al. X-linked Alport syndrome caused by splicing mutations in COL4A5. Clin J Am Soc Nephrol 2014;9:1958-64. [Crossref] [PubMed]
- Horinouchi T, Nozu K, Yamamura T, et al. Detection of Splicing Abnormalities and Genotype-Phenotype Correlation in X-linked Alport Syndrome. J Am Soc Nephrol 2018;29:2244-54. [Crossref] [PubMed]
- Hashimura Y, Nozu K, Kaito H, et al. Milder clinical aspects of X-linked Alport syndrome in men positive for the collagen IV α5 chain. Kidney Int 2014;85:1208-13. [Crossref] [PubMed]
- James CC, Smyth JW. Alternative mechanisms of translation initiation: An emerging dynamic regulator of the proteome in health and disease. Life Sci 2018;212:138-44. [Crossref] [PubMed]
- Nozu K, Nakanishi K, Abe Y, et al. A review of clinical characteristics and genetic backgrounds in Alport syndrome. Clin Exp Nephrol 2019;23:158-68. [Crossref] [PubMed]
- Ding F, Wickman L, Wang SQ, et al. Accelerated podocyte detachment and progressive podocyte loss from glomeruli with age in Alport Syndrome. Kidney Int. 2017;92:1515-25. [Crossref] [PubMed]
- Haas M. Thin glomerular basement membrane nephropathy: incidence in 3471 consecutive renal biopsies examined by electron microscopy. Arch Pathol Lab Med 2006;130:699-706. [Crossref] [PubMed]
- Malone AF, Phelan PJ, Hall G, et al. Rare hereditary COL4A3/COL4A4 variants may be mistaken for familial focal segmental glomerulosclerosis. Kidney Int 2014;86:1253-9. [Crossref] [PubMed]
- Wu Y, Hu P, Xu H, et al. A novel heterozygous COL4A4 missense mutation in a Chinese family with focal segmental glomerulosclerosis. J Cell Mol Med 2016;20:2328-32. [Crossref] [PubMed]
- Fu XJ, Nozu K, Eguchi A, et al. X-linked Alport syndrome associated with a synonymous p.Gly292Gly mutation alters the splicing donor site of the type IV collagen alpha chain 5 gene. Clin Exp Nephrol 2016;20:699-702. [Crossref] [PubMed]
- Migeon BR. X inactivation, female mosaicism, and sex differences in renal diseases. J Am Soc Nephrol 2008;19:2052-9. [Crossref] [PubMed]
- Temme J, Peters F, Lange K, et al. Incidence of renal failure and nephroprotection by RAAS inhibition in heterozygous carriers of X-chromosomal and autosomal recessive Alport mutations. Kidney Int 2012;81:779-83. [Crossref] [PubMed]
- Malone AF, Funk SD, Alhamad T, et al. Functional assessment of a novel COL4A5 splice region variant and immunostaining of plucked hair follicles as an alternative method of diagnosis in X-linked Alport syndrome. Pediatr Nephrol 2017;32:997-1003. [Crossref] [PubMed]
- Savige J, Colville D, Rheault M, et al. Alport Syndrome in Women and Girls. Clin J Am Soc Nephrol 2016;11:1713-20. [Crossref] [PubMed]