
Novel nucleic acid aptamer gold (Au)-nanoparticles (AuNPs-AptHLA-G5-1 and AuNPs-AptHLA-G5-2) to detect the soluble human leukocyte antigen G5 subtype (HLA-G5) in liquid samples
Introduction
Human leukocyte antigen G5 subtype (HLA-G5) is an essential human histocompatibility complex [major histocompatibility complex (MHC)], which belongs to the Ib subtype of MHC (1-3). As an important regulatory factor of the immune system, HLA-G5 can alleviate the damage caused by verification, alleviate the rejection caused by the suppression of homologous and allologous immune-rejection, and play a protective role in the body (4-6). It is essential that HLA-G5 is selectively expressed in maternal-foetal interface tissues, where it is mainly distributed in the extravillous cytotrophoblast, and can play a protective role during embryonic development and pregnancy (7-9). In in vitro fertilization (IVF) embryo culture medium, human embryos can secrete soluble HLA-G protein (soluble HLA-G5, sHLA-G5) at the 4–8-cell stage, and this secretion is closely related to the successful implantation of in vitro fertilized embryos (10,11). Therefore, it is critical to detect HLA-G5, especially HLA-G5 expression in embryo fluid, during in vitro embryo incubation and culture. However, in commonly used embryo culture medium (approximately 12 µL), the specificity and sensitivity of traditional ELISA methods to detect sHLA-G5 are insufficient. Additionally, specific limitations, such as cumbersome operation and multiple steps, hamper its use for efficient detection. Therefore, exploring the ligands that can recognize sHLA-G5 with high specificity and high affinity is of great importance to improve the specificity and sensitivity of its detection.
Nucleic acid aptamers (aptamers) are single-stranded oligonucleotides that can specifically bind to proteins or other biological macromolecules obtained from a random single-stranded oligonucleotide library (12-14). Aptamers are designed with colloidal gold nanotechnology, making them a portable, fast, simple, and highly-sensitive detection method (15,16). Gold nanoparticles were incubated with aptamers screened in this study to obtain aptamer nano-gold detection reagents (AuNPs-AptHLA-G5-1 and AuNPs-AptHLA-G5-2). When HLA-G5 is present in a liquid test sample, it competitively binds to the aptamer in the detection reagent, causing the aptamer to dissociate from the gold nanoparticles. The use of novel AuNPs to detect HLA-G5 will assist both IVF research and the broader field of diagnostic and therapeutic procedures in obstetrics and gynaecology. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3334).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the ethics committee of the First Medical Center of Chinese PLA General Hospital (approval No.: s2019-196-01). The informed consent was waived due to the anonymous data and non-intervention feature of the study.
Materials and reagents
All the reagents used in the procedure of prokaryotic expression and purification of HLA-G5 protein were produced by the BBI Life Sciences Corporation (Shanghai, China) including the LB Broth Agar Medium, Rosetta competent bacteria, protein expression inducer, non-pre-stained protein marker, pre-stained protein marker, TMB chromogenic Kit, Western Blot primary antibody, Western Blot secondary antibody, and the protein concentration quantitative Kit. Reagents used for the ligand system evolution and aptamer screening included Dynabeads MyOneTM Carboxylic Acid (Thermo, USA), DPBS (BBI, USA), BSA (BBI, USA), NHS (Sigma, USA), EDC (J&K Scientific, China), Pfu DNA polymerase (BBI, USA), 2X high fidelity PCR mix premix (BBI, USA), sodium acetate (BBI Life Sciences Corporation, China), ethanolamine (BBI Life Sciences Corporation, China), TEMED (BBI Life Sciences Corporation, China), and dialysis bag 3.5KD (BBI Life Sciences Corporation, China).
Prokaryotic expression system of HLA-G
The pET28a vector containing the full-length region of HLA-G5 was cloned into a recombinant plasmid and transduced to Rosetta (DE3) Escherichia coli (E. coli), which was then cultured. The culture medium was continuously tested during the shaking process, and when the O.D. value at 600 nm reached 0.6, the inducer IPTG (Isopropyl-beta-D-thiogalactopyranoside) (100 mmol/L) was added, and the culture was continued at 20 °C for 16 h. The bacteria were collected by centrifugation then lysed, and the samples were prepared for SDS-PAGE. The separation and purification of HLA-G5 was performed using Ni-NTA (Ni column affinity chromatography packing) affinity chromatography methods (17,18), and the purified HLA-G5 protein was analyzed using SDS-PAGE.
SELEX methods to prepare the aptamer of HLA-G5
The aptamer of HLA-G5 was screened from a DNA library using methods from the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) (19-22). Briefly, the carboxyl magnetic beads were incubated with HLA-G5 protein (concentration of 0.35 mg/mL) for 40 min on a shaker at room temperature. The DNA library was then diluted and distributed into PCR tubes for denaturation, and mixed with carboxyl magnetic beads combined with HLA-G5. Lastly, the carboxyl magnetic beads were separated from the system by centrifugation. The potential aptamer (the ssDNA) with HLA-G5 from the DNA library was identified using qPCR.
Detection of HLA-G based on AuNPs-aptamer sensing method
Gold nanoparticles (AuNPs) were synthesized by citrate reduction following the methods described by Zhang [2021] and Xia [2020] (23,24). A 2% chloroauric acid solution (200 mL) was boiled under stirring conditions, and 6 mL of 2% trisodium citrate solution was quickly added. The solution was kept heated for 15 min under stirring conditions until the colour became wine red, and the prepared AuNPs solution was then stored at 4 °C and protected from light. Transmission electron microscope (TEM) was used to examine the prepared gold nanoparticles (25,26). A concentration gradient for HLA-G was prepared using a standard solution (10, 20, 50, 100, 200, 500, and 1,000 nmol/L) and stored at 4 °C for later use. An incubation buffer (Na2HPO4-KH2PO4, pH 7.6, 50 mmol/L Na+, 5 mmol/L K+, 5 mmol/L Mg2+, 1 mmol/L Ca2+) was used to dilute HLA-G5-Apt1 and HLA-G5-Apt2, and 50 µL AuNPs solution was mixed with a 4 µmol/L aptamer solution (25 µL) at room temperature for 5 min. About 25 µL of HLA-G5 standard solution with a concentration gradient was then incubated at room temperature for 20 min, and at the end of the reaction, 10 µL of NaCl solution (900 mmol/L) was added and mixed well. Colour changes were observed by absorbance changes at 520 and 650 nm were measured using a microplate reader.
Statistical analysis
The continuous variables are expressed as mean ± standard deviation (SD). The results come from three replicates with similar trends were analyzed by using the linear regression analysis. All the statistical analyses were performed by using the IBM® SPSS® Statistics software Version 22.
Results
HLA-G proteins obtained from the prokaryotic expression system
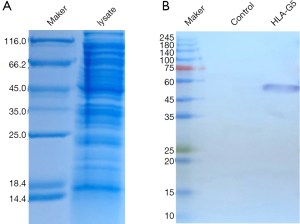
HLA-G5 proteins were obtained using the prokaryotic expression system, and successfully induced HLA-G expression in E. coli (Figure 1). The results of Coomassie brilliant blue staining showed that the HLA-G band was visible, and the molecular weight of the band was correct. Images of bacterial lysate and the purification of HLA-G protein are shown in Figure 1A,1B, respectively.
Aptamers of HLA-G5 with high affinity
The target protein (HLA-G5) was successfully coupled to the protein detection chip, and a surface plasmon resonance (SPR) test was performed. Affinity binding of HLA-G5 protein to the aptamer [HLA-G5-1 (>Seq2_778_289773_0.0027, TGTTGACCAGGGACTGCTCGGGATTGCG GATGTCAA) or HLA-G5-2 (>Seq5_368_289773_0.0013, TGTTGACCAGGGACTGCTCGGGA TTGCGGACTTCAA)] is shown in the SPR curves (Figure 2), and the single curve shown above fits the KD value (affinity). The value corresponding to KD below Figure 2 is the affinity dissociation constant. For HLA-G5-Apt1 (AptHLA-G5-1 =6.356×10−7 µmol/L), and for HLA-G5-Apt2 (AptHLA-G5-2 =2.949×10−7 µmol/L). This demonstrates that HLA-G5 aptamers with high affinity were successfully obtained.

Use of Au-nano-particles to examine HLA-G5
To verify the accuracy and reliability of the HLA-G detection method using nano-gold-aptamer colorimetric sensing, the aptamer and nano-Au were mixed to form the novel aptamer Au-nanoparticles AuNPs-AptHLA-G1 and AuNPs-AptHLA-G2 (Figure 3), and transmission electron microscopy show each had a uniform particle size of about 10 nm (Figure 4). HLA-G5 was detected by AuNPs-AptHLA-G5-1 and AuNPs-AptHLA-G5-2 using a colorimetric sensing method and in liquid samples (Figure 5). The AuNPs-AptHLA-G5-1 colorimetric sensing method had a detection limit of approximately 20 ng/mL (recovery range: 98.7% to 102.0%) while that of AuNPs-AptHLA-G5-2 was as low as 20 ng/mL (recovery range: 98.9% to 103.6%) (Figure 5). The standard-curves of AuNPs-AptHLA-G5-1 and AuNPs-AptHLA-G5-2 obtained by incubation with HLA-G5 are shown in Figure 5 and Table 1.
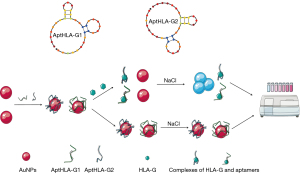
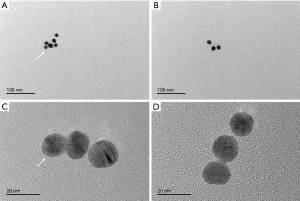
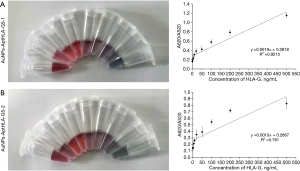
Table 1
| Au-nano particles | HLA-G5 (ng/mL) | Testing results (ng/mL) | Recovery rate (%) | Recovery rate SD (%, n=3) |
|---|---|---|---|---|
| AuNPs-AptHLA-G5-1 | 20 | 20.24±1.21 | 101.2 | 5.9 |
| 50 | 49.35±3.16 | 98.7 | 6.4 | |
| 100 | 99.80±9.02 | 99.8 | 9 | |
| 200 | 204.00±15.32 | 102 | 7.4 | |
| AuNPs-AptHLA-G5-2 | 20 | 20.72±1.13 | 103.6 | 5.4 |
| 50 | 50.70±3.22 | 101.4 | 6.3 | |
| 100 | 98.90±6.15 | 98.9 | 6.2 | |
| 200 | 198.00±10.28 | 99 | 5.2 |
HLA-G5, human leukocyte antigen G5 subtype.
Discussion
With the influence of many factors such as social structure, population aging, environmental pollution, late marriage, and late childbirth, countries worldwide have generally seen a decline in marriage and fertility rates and a gradual increase in the incidence of fertility disorders (26,27). In addition, the gradual implementation of the Chinese governments comprehensive liberalization of the second-child policy after 2016 has seen some families who may have lost their only child or had only one child with genetic diseases or defects, to attempt an additional pregnancy using strategies such as assisted reproduction. The most effective assisted reproduction technology at present remains in vitro fertilization (IVF), which is mainly used in cases of (I) fallopian tube disease (tubal blockage caused by pelvic inflammatory disease); (II) endometriosis and other endometrial diseases; (III) male oligospermia, asthenospermia, or teratospermia; and (IV) conditions where there is a high risk of genetic diseases or genetic defects (28,29). Despite advancements in IVF-related technology for diagnosis and treatment (to the third generation), its success rate is affected by the success rate of embryo transfer and the survival rate of foetus delivery, and remains low (approximately 20%) (30,31). For this reason, researchers are eager to develop new strategies to improve the IVF success rate and determine indicators of success. Studies have shown that single-cell omics research before embryo transfer can help solve this problem (28,32,33), but this technique requires the extraction of a small number of cells and tissues from the embryo, which may damage it (28,32,33). Since multiple pregnancies are unfavourable to the health of mothers and infants, choosing a single embryo for transplantation without reducing the pregnancy rate can effectively prevent their occurrence (34,35). However, the challenge plaguing researchers and first-line clinicians has always been selecting embryos that can better implant and survive throughout the pregnancy. At present, a widely used strategy in clinical diagnosis and treatment is to select embryos for transfer based on embryo morphology scores (36,37). The outstanding problem of this strategy is that it is impossible to establish a complete standard and quantitatively study embryo morphology, which may not be wholly consistent with its developmental potential. For this reason, it is of great significance to use embryonic fluid to establish a completely non-invasive, efficient, convenient, and sensitive detection method.
Distinct from the most classic MHC type Ia molecules, HLA-G5 belongs to the non-classical MHC complex Ib subtype. As an essential immune tolerance regulator in the human body, HLA-G5 is involved in processes such as foetal tolerance and autoimmunity during pregnancy, as well as inflammatory diseases and receiving allotransplantation from patients (37,38), and can achieve immune tolerance and protect the human body by interacting with inhibitory receptors on the surface of immune cells. HLA-G can interact with immunoglobulin-like proteins on dendritic cells (DC) (39,40), and through such self-made interactions, can suppress immune cells (41). Importantly, in normal tissues, HLA-G is most abundantly distributed on the surface of trophoblast cells in the placenta, which can effectively inhibit the local immune response in the uterus and promote the tolerance of the mother to the foetus (6,42). Based on this characteristic, the expression level of HLA-G is closely related to the success rate of embryo transfer during IVF treatment. In this study, an HLA-G aptamer was successfully prepared, which laid the foundation for detecting HLA-G expression in a minimal volume of embryonic fluid.
Biological macromolecules, including DNA, RNA, antibodies, and proteins, can be used as targets for aptamer detection (43). Traditional research mostly uses western blot, southern blot, or northern blot to detect biological macromolecules, and for soluble proteins in solution, ELISA is widely used (44,45). DNA aptamers are used for direct generation using a synthetic DNA library by the SELEX method. Aptamers are not only widely used in clinical testing, but their non-immunogenicity also provides them with significant therapeutic potential (46). Aptamers with very high affinity to the target molecule help in early-stage diagnosis during disease progression, patient treatment, and the prevention of further disease spread. High-affinity aptamers are currently being tested in various clinical trials, and because of the high adaptability of chemically modified aptamers, stable aptamers have been produced and have been widely used in these trials (47). The preparation of HLA-G aptamers in this study is not only helpful for HLA-G-related research but also expands our broader understanding of aptamers. them
Traditional detection methods use chemiluminescence methods such as horseradish peroxidase (HRP), which has limited amplification of trace signals. Combining immunoassay technology with aptamers with golden nanoparticles or microfluidic chip technology, can build a portable and straightforward chip device to establish a fast, simple, and highly-sensitive detection method. Additionally, the radioisotopes (31P) can incorporate aptamer nucleic acid molecules to amplify the signal (48).
Acknowledgments
Funding: This work was supported by the National Key Research and Development Program of China (No. 2018YFC1003100).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3334
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-3334
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3334). The authors report funding support from the National Key Research and Development Program of China (No. 2018YFC1003100). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the ethics committee of the First Medical Center of Chinese PLA General Hospital (approval No.: s2019-196-01). The informed consent was waived due to the anonymous data and non-intervention feature of the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Talotta R, Atzeni F, Sarzi-Puttini P, et al. Psoriatic arthritis: From pathogenesis to pharmacologic management. Pharmacol Res 2019;148:104394 [Crossref] [PubMed]
- Ye BJ, Lee HH, Yoo EJ, et al. TonEBP in dendritic cells mediates pro-inflammatory maturation and Th1/Th17 responses. Cell Death Dis 2020;11:421. [Crossref] [PubMed]
- Liu L, Wang L, Zhao L, et al. The Role of HLA-G in Tumor Escape: Manipulating the Phenotype and Function of Immune Cells. Front Oncol 2020;10:597468 [Crossref] [PubMed]
- Zidi I. Puzzling out the COVID-19: Therapy targeting HLA-G and HLA-E. Hum Immunol 2020;81:697-701. [Crossref] [PubMed]
- Rashidi S, Farhadi L, Ghasemi F, et al. The potential role of HLA-G in the pathogenesis of HBV infection: Immunosuppressive or immunoprotective? Infect Genet Evol 2020;85:104580 [Crossref] [PubMed]
- Papúchová H, Meissner TB, Li Q, et al. The Dual Role of HLA-C in Tolerance and Immunity at the Maternal-Fetal Interface. Front Immunol 2019;10:2730. [Crossref] [PubMed]
- Persson G, Jørgensen N, Nilsson LL, et al. A role for both HLA-F and HLA-G in reproduction and during pregnancy? Hum Immunol 2020;81:127-33. [Crossref] [PubMed]
- Lee SJ, Kim J, Choi WH, et al. Immunophenotype and Immune-Modulatory Activities of Human Fetal Cartilage-Derived Progenitor Cells. Cell Transplant 2019;28:932-42. [Crossref] [PubMed]
- da Silva IL, Montero-Montero L, Ferreira E, et al. New Insights Into the Role of Qa-2 and HLA-G Non-classical MHC-I Complexes in Malignancy. Front Immunol 2018;9:2894. [Crossref] [PubMed]
- Lin A, Yan WH. Heterogeneity of HLA-G Expression in Cancers: Facing the Challenges. Front Immunol 2018;9:2164. [Crossref] [PubMed]
- Kametani Y, Shiina T, Suzuki R, et al. Comparative immunity of antigen recognition, differentiation, and other functional molecules: similarities and differences among common marmosets, humans, and mice. Exp Anim 2018;67:301-12. [Crossref] [PubMed]
- Engelberg S, Netzer E, Assaraf YG, et al. Selective eradication of human non-small cell lung cancer cells using aptamer-decorated nanoparticles harboring a cytotoxic drug cargo. Cell Death Dis 2019;10:702. [Crossref] [PubMed]
- Liu J, Guo B. RNA-based therapeutics for colorectal cancer: Updates and future directions. Pharmacol Res 2020;152:104550 [Crossref] [PubMed]
- Moradi A, Pourseif MM, Jafari B, et al. Nanobody-based therapeutics against colorectal cancer: Precision therapies based on the personal mutanome profile and tumor neoantigens. Pharmacol Res 2020;156:104790 [Crossref] [PubMed]
- Zhang K, Li H, Wang W, et al. Application of Multiplexed Aptasensors in Food Contaminants Detection. ACS Sens 2020;5:3721-38. [Crossref] [PubMed]
- Girotti A, Escalera-Anzola S, Alonso-Sampedro I, et al. Aptamer-Functionalized Natural Protein-Based Polymers as Innovative Biomaterials. Pharmaceutics 2020;12:1115. [Crossref] [PubMed]
- Matsuo S, Hiasa M, Omote H. Functional characterization and tissue localization of the facilitative glucose transporter GLUT12. J Biochem 2020;168:611-20. [Crossref] [PubMed]
- Kielkopf CL, Bauer W, Urbatsch IL. Purification of Polyhistidine-Tagged Proteins by Immobilized Metal Affinity Chromatography. Cold Spring Harb Protoc 2020;2020:102194 [Crossref] [PubMed]
- Zhao J, Wang M, Yang Y, et al. CD123 thioaptamer protects against sepsis via the blockade between IL-3/CD123 in a cecal ligation and puncture rat model. Nucleosides Nucleotides Nucleic Acids 2021;40:16-31. [Crossref] [PubMed]
- Wang M, Wu H, Li Q, et al. Novel Aptamer-Functionalized Nanoparticles Enhances Bone Defect Repair By Improving Stem Cell Recruitment. Int J Nanomedicine 2019;14:8707-24. [Crossref] [PubMed]
- Wang M, Wu H, Duan M, et al. SS30, a novel thioaptamer targeting CD123, inhibits the growth of acute myeloid leukemia cells. Life Sci 2019;232:116663 [Crossref] [PubMed]
- Song M, Li G, Zhang Q, et al. De novo post-SELEX optimization of a G-quadruplex DNA aptamer binding to marine toxin gonyautoxin 1/4. Comput Struct Biotechnol J 2020;18:3425-33. [Crossref] [PubMed]
- Zhang D, Liu Y, Ding J, et al. Label-free colorimetric assay for arsenic(III) determination based on a truncated short ssDNA and gold nanoparticles. Mikrochim Acta 2021;188:38. [Crossref] [PubMed]
- Xia X, Li M, Wang M, et al. Development of Ochratoxin A Aptasensor Based on Au Nanoparticles@g-C3N4. J Biomed Nanotechnol 2020;16:1296-303. [Crossref] [PubMed]
- Tan JH, Cao RC, Zhou L, et al. EMC6 regulates acinar apoptosis via APAF1 in acute and chronic pancreatitis. Cell Death Dis 2020;11:966. [Crossref] [PubMed]
- Zhao X, Liu Y, Zhang A, et al. Logistic regression analyses of factors affecting fertility of intrauterine adhesions patients. Ann Transl Med 2020;8:49. [Crossref] [PubMed]
- Hussain T, Tan B, Murtaza G, et al. Role of Dietary Amino Acids and Nutrient Sensing System in Pregnancy Associated Disorders. Front Pharmacol 2020;11:586979 [Crossref] [PubMed]
- Liu S, Xie Y, Li F, et al. Effectiveness of ultra-long protocol on in vitro fertilization/intracytoplasmic sperm injection-embryo transfer outcome in infertile women with endometriosis: A systematic review and meta-analysis of randomized controlled trials. J Obstet Gynaecol Res 2021;47:1232-42. [Crossref] [PubMed]
- Wei D, Liu JY, Sun Y, et al. Frozen versus fresh single blastocyst transfer in ovulatory women: a multicentre, randomised controlled trial. Lancet 2019;393:1310-8. [Crossref] [PubMed]
- Gleicher N, Barad DH. Assessing in-vitro fertilisation at age 40 years. Lancet 2019;393:1181-3. [Crossref] [PubMed]
- Ejzenberg D, Andraus W, Baratelli Carelli Mendes LR, et al. Livebirth after uterus transplantation from a deceased donor in a recipient with uterine infertility. Lancet 2019;392:2697-704. [Crossref] [PubMed]
- Li L, Guo F, Gao Y, et al. Single-cell multi-omics sequencing of human early embryos. Nat Cell Biol 2018;20:847-58. [Crossref] [PubMed]
- Oliveira CS, Romano Quintão CC, de Freitas C, et al. Post implantation development reveals that biopsy procedure can segregate "healthy" from "unhealthy" bovine embryos and prevent miscarriages. Anim Reprod Sci 2017;184:51-8. [Crossref] [PubMed]
- Gleicher N, Kuhnir VA, Barad DH. Unexplained infertility. Lancet 2018;392:1516-7. [Crossref] [PubMed]
- Farquhar CM, Liu E, Armstrong S, et al. Intrauterine insemination with ovarian stimulation versus expectant management for unexplained infertility (TUI): a pragmatic, open-label, randomised, controlled, two-centre trial. Lancet 2018;391:441-50. [Crossref] [PubMed]
- Zaninovic N, Rosenwaks Z. Artificial intelligence in human in vitro fertilization and embryology. Fertil Steril 2020;114:914-20. [Crossref] [PubMed]
- Griesinger G, Blockeel C, Kahler E, et al. Dydrogesterone as an oral alternative to vaginal progesterone for IVF luteal phase support: A systematic review and individual participant data meta-analysis. PLoS One 2020;15:e0241044 [Crossref] [PubMed]
- Xu X, Zhou Y, Wei H. Roles of HLA-G in the Maternal-Fetal Immune Microenvironment. Front Immunol 2020;11:592010 [Crossref] [PubMed]
- Ye Q, Sung TC, Yang JM, et al. Generation of universal and hypoimmunogenic human pluripotent stem cells. Cell Prolif 2020;53:e12946 [Crossref] [PubMed]
- Contini P, Murdaca G, Puppo F, et al. HLA-G Expressing Immune Cells in Immune Mediated Diseases. Front Immunol 2020;11:1613. [Crossref] [PubMed]
- Bai Y, Liang J, Liu W, et al. Possible roles of HLA-G regulating immune cells in pregnancy and endometrial diseases via KIR2DL4. J Reprod Immunol 2020;142:103176 [Crossref] [PubMed]
- Xu HH, Yan WH, Lin A. The Role of HLA-G in Human Papillomavirus Infections and Cervical Carcinogenesis. Front Immunol 2020;11:1349. [Crossref] [PubMed]
- Alkhamis O, Canoura J, Yu H, et al. Innovative engineering and sensing strategies for aptamer-based small-molecule detection. Trends Analyt Chem 2019;121:115699 [Crossref] [PubMed]
- Yuan S, Chan JFW, Chik KKH, et al. Discovery of the FDA-approved drugs bexarotene, cetilistat, diiodohydroxyquinoline, and abiraterone as potential COVID-19 treatments with a robust two-tier screening system. Pharmacol Res 2020;159:104960 [Crossref] [PubMed]
- Di Desidero T, Orlandi P, Gentile D, et al. Pharmacological effects of vinorelbine in combination with lenvatinib in anaplastic thyroid cancer. Pharmacol Res 2020;158:104920 [Crossref] [PubMed]
- Bing T, Zhang N, Shangguan D. Cell-SELEX, an Effective Way to the Discovery of Biomarkers and Unexpected Molecular Events. Adv Biosyst 2019;3:e1900193 [Crossref] [PubMed]
- Ozer T, Geiss BJ, Henry CS. Review-Chemical and Biological Sensors for Viral Detection. J Electrochem Soc 2020;167:037523 [Crossref] [PubMed]
- Filippi L, Bagni O, Nervi C. Aptamer-based technology for radionuclide targeted imaging and therapy: a promising weapon against cancer. Expert Rev Med Devices 2020;17:751-8. [Crossref] [PubMed]
(English Language Editor: B. Draper)


