
Olaparib effectively treats local recurrence of extrahepatic cholangiocarcinoma in a patient harboring a BRCA2-inactivating mutation: a case report
Introduction
Cholangiocarcinoma (CCA), stratified anatomically into intrahepatic CCA (ICC) and extrahepatic CCA (ECC), is a malignant tumor with poor prognosis and high recurrence rate (1). CCA is infrequently observed in Europe and the USA; however, it is more common in China (>6/100,000 cases) (2). Surgery remains the primary treatment option for biliary tract cancers; however, it is considered unsuitable for more than two-thirds of CCA patients (3). There are few chemotherapeutic options that provide only limited benefits for advanced patients according to the National Comprehensive Cancer Network (NCCN) clinical practice guidelines. Given the emerging evidence regarding the actionable targets for treating CCA, molecular testing of unresectable or metastatic tumors is recommended (4). At present, targeted therapies, including larotrectinib and entrectinib for NTRK gene fusion-positive tumors (5,6), and pembrolizumab for MSI-H/dMMR tumors are recommend for advanced CCA (7). As pemigatinib has great therapeutic potential in previously treated patients with CCA who have FGFR2 fusions or rearrangements and was accelerated approved by the US Food and Drug Administration (FDA) (8), and ivosidenib was demonstrated efficacy and safety in advanced, IDH1-mutant CCA (9), these 2 target therapies are recommended for the treatment of CCA in the latest NCCN guidelines.
BRCA2 inactivation plays an important role in hepatocellular carcinoma development and progression, and patients with mutant BRCA2 alleles are more likely to develop malignant liver tumors (10). BRCA2-inactivating mutation frequency in ECC is approximately 4% (11). Few reports of BRCA2-inactivating gene rearrangements exist for bile duct carcinoma. In the present study, we reported a case of a postoperative recurrence ECC patient harboring a BRCA2-inactivating rearrangement mutation that had an obvious reaction to olaparib therapy. We present the following article in accordance with the CARE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3681).
Case presentation
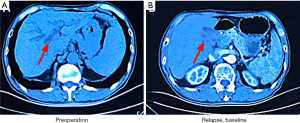
In June 2015, a 68-year-old man presented yellow discoloration of the skin without an obvious etiology, accompanied by nausea and bloating. He did not receive any treatment initially, and the skin discoloration gradually became more pronounced. In August 2015, the patient was treated at Liaoning Cancer Hospital and Institute, Shenyang, China. Abdominal computed tomography (CT) revealed intrahepatic bile duct dilatation, obstruction at the hepatic hilar region proximal to the common hepatic duct, and splenomegaly (Figure 1A). The patient underwent radical surgical resection for CCA. Postoperative pathological assessment revealed a moderately differentiated adenocarcinoma (1.5-cm diameter) arising from the bile duct and infiltrating the serosa, nerve involvement, and vascular tumor thrombus. Immunohistochemistry showed that the tumor comprised CK7+, CK20–, P53+ (~90%), and Ki-67+ (~90%) cells; ECC without metastases was diagnosed. The patient refused adjuvant therapy after surgery due to personal reasons. In February 2017, abdominal CT revealed local recurrence (Figure 1B). The patient refused chemotherapy because he was not experiencing discomfort.
In May 2017, surgically resected tissues were examined for 450 tumor-associated genes by OrigiMed (Shanghai, China) (Table 1) (2). Nine tumor-related gene variants were detected by next-generation sequencing, including a BRCA2 rearrangement, cyclin-dependent kinase inhibitor 2A (CDKN2A) P81L, tumor protein p53 (TP53) Y163C, and ataxia-telangiectasia and rad3 related (ATR) K1557Sfs*8 mutations, as well as kirsten rat sarcoma (KRAS), C-X-C chemokine receptor type 4 (CXCR4), myeloid cell leukemia 1(MCL1), Fos proto-oncogene (FOS), and Jun proto-oncogene (JUN) amplifications.. BRCA2 rearrangement was observed between chromosome 20 p11.21 and exons 21–27 of BRCA2 (Figure 2), and might have had deletions of exons 1–20, thereby impairing BRCA2 function.
Table 1
| Gene | Mutation | Variation frequency |
|---|---|---|
|
|
P81L | 35% |
|
|
Rearrangement | – |
|
|
Amplification | – |
|
|
Y163C | 31% |
|
|
K1557Sfs*8 | 3% |
|
|
Amplification | – |
|
|
Amplification | – |
|
|
Amplification | – |
|
|
Amplification | – |
*, stands for termination codon.
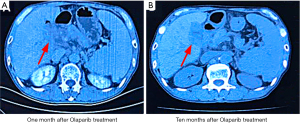
In October 2017, the patient complained of upper abdominal pain and refused radiotherapy or chemotherapy. Oral olaparib administration was initiated. Efficacy evaluation 1 month after treatment confirmed stable disease (Figure 3A). Until August 2018, the patient continued to benefit from olaparib treatment, and the disease remained stable without significant adverse reactions, such as anemia, vomiting, diarrhea, infection, or fatigue (Figure 3B) (12). The patient achieved progression-free survival (PFS) of >10 months, exceeding the 8-month median PFS obtained with the gemcitabine + cisplatin regimen recommended by the NCCN (13). During the first 10 months of taking olaparib, the patient was followed closely. However, the patient then returned to his hometown and we were unable to conduct further treatment or obtain imaging data. In January 2020, we were informed that the patient had died, with an overall survival (OS) of 27 months.
This case report shows that genetic testing for patients with biliary tract cancer, and olaparib treatment of patients with ECC harboring BRCA2 rearrangements, are likely to be beneficial in clinical settings.
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Discussion
BRCA1 and BRCA2 are tumor-suppressor genes and play important roles in homologous recombination-mediated, double-stranded DNA break repair, thereby maintaining genomic stability. The mutant status of these genes can increase the risk of breast, ovarian, and gastric cancers. The FDA has approved the use of poly (ADP-ribose) polymerase (PARP) inhibitors, olaparib, talazoparib, rucaparib, and niraparib, for patients with breast and ovarian cancers involving BRCA mutations, and platinum-sensitive pancreatic and epithelial ovarian, fallopian tube, or primary peritoneal cancers.
Tumor cells with homologous recombination defects due to BRCA loss of function are more dependent on PARP-mediated DNA repair. Preclinical studies have demonstrated that PARP inhibitors can kill homologous recombination-defective tumor cells through a synthetic lethal mechanism (14,15). Among small-cell lung cancer patients with a BRCA1/2 mutation (n=23) who received PARP inhibitor (talazoparib) monotherapy, 2 patients achieved partial remission and 4 achieved disease stabilization (16). Additionally, a refractory ICC patient with a heterozygous germline BRCA2 p.S2670* mutation achieved partial response from olaparib monotherapy (17). The clinical effectiveness of PARP inhibitors for treating CCA patients with BRCA somatic mutations remains unclear. In the present study, we first reported the clinical benefits of olaparib in an ECC patient harboring a BRCA2-inactivating rearrangement.
The tumor suppressor gene, CDKN2A, encodes 2 unrelated proteins, p16INK4a and p14ARF (18). p16INK4a or p14ARF loss of function through mutations in CDKN2A, or via promoter hypermethylation, disturbs Rb/CDK and p53 signaling, resulting in uncontrolled cell growth and cancer (19). CDKN2A P81L causes a proline-to-leucine substitution at position 81 of p16INK4a. In vitro experiments have shown that the P81L mutation reduces the inhibitory effect of p16INK4a, prevents it from binding to CDK4 and inhibits protein activity. The mutant p16INK4a binds to CDK6 instead (20). A structural analysis showed that the mutant protein has folding defects (21).
There are no FDA-approved anticancer drugs targeting CDKN2A (p16INK4a). However, given that CDKN2A (p16INK4a) loss leads to CDK4/6 activation and cell proliferation, CDK inhibitors may represent a therapeutic option for cancer patients with CDKN2A loss-of-function variants. The FDA has approved the CDK4/6 inhibitors palbociclib, ribociclib, and abemaciclib for the treatment of estrogen receptor-positive, HER2-negative, advanced breast cancer. Because the level of evidence between CDK4/6 inhibitors and molecular markers is lower compared with PARP inhibitors, CDK 4/6 inhibitors were not considered in this case.
TP53 encodes transcription factor p53 and is the most frequently mutated tumor-suppressor gene in human malignancies (22). In vitro experiments have shown that Y163C mutations decrease the transactivation capacity of p53, rendering the protein unable to suppress cell proliferation and induce apoptosis (23).
The DNA damage repair gene, ATR, encodes a serine/threonine kinase. The ATR K1557Sfs*8 mutation causes a lysine-to-serine conversion at position 1557, and the 8th codon is terminated in the new reading frame. Based on the location of the premature termination codon, it is predicted that this mutation leads to nonsense mutation-mediated mRNA degradation, and therefore, a lack of ATR protein.
RAS oncogenes, NRAS, KRAS, and HRAS, are commonly mutated in human tumors. KRAS amplification is observed in various tumors and is associated with increased levels of KRAS protein (24,25), which can in turn cause malignant transformation of cells through the activation of mitogen-activated protein kinase/extracellular signal-regulated kinase signaling (26). Unfortunately, there are no FDA-approved anticancer drugs targeting TP53, ATR, or KRAS. There are also no drugs targeting CXCR4, MCL1, FOS, and JUN amplifications in cancer.
In the present study, we reported on a patient with ECC harboring a BRCA2-inactivating rearrangement mutation responded to olaparib therapy with a PFS of >10 months and an OS of 27 months. This is the first report demonstrating the clinical benefits of olaparib in a patient with ECC harboring a BRCA2 rearrangement. This case also highlights the importance of next-generation sequencing for providing valuable information for designing effective targeted therapies for ECC. Evidence-based decisions help determine the best customized treatment option.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3681
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3681). JS, ML, and MX are from Shanghai OrigiMed Co. Ltd. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fléjou JF. WHO Classification of digestive tumors: the fourth edition. Ann Pathol 2011;31:S27-31.
- Banales JM, Cardinale V, Carpino G, et al. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol 2016;13:261-80. [Crossref] [PubMed]
- Yamamoto M, Takasaki K, Yoshikawa T. Lymph node metastasis in intrahepatic cholangiocarcinoma. Jpn J Clin Oncol 1999;29:147-50. [Crossref] [PubMed]
- Lowery MA, Ptashkin R, Jordan E, et al. Comprehensive Molecular Profiling of Intrahepatic and Extrahepatic Cholangiocarcinomas: Potential Targets for Intervention. Clin Cancer Res 2018;24:4154-61. [Crossref] [PubMed]
- Scott LJ. Larotrectinib: First Global Approval. Drugs 2019;79:201-6. [Crossref] [PubMed]
- Doebele RC, Drilon A, Paz-Ares L, et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1-2 trials. Lancet Oncol 2020;21:271-82. [Crossref] [PubMed]
- Marabelle A, Le DT, Ascierto PA, et al. Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J Clin Oncol 2020;38:1-10. [Crossref] [PubMed]
- Abou-Alfa GK, Sahai V, Hollebecque A, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol 2020;21:671-84. [Crossref] [PubMed]
- Abou-Alfa GK, Macarulla T, Javle MM, et al. Ivosidenib in IDH1-mutant, chemotherapy-refractory cholangiocarcinoma (ClarIDHy): a multicentre, randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol 2020;21:796-807. [Crossref] [PubMed]
- Katagiri T, Nakamura Y, Miki Y. Mutations in the BRCA2 gene in hepatocellular carcinomas. Cancer Res 1996;56:4575-7. [PubMed]
- Nakamura H, Arai Y, Totoki Y, et al. Genomic spectra of biliary tract cancer. Nat Genet 2015;47:1003-10. [Crossref] [PubMed]
- Ledermann J, Harter P, Gourley C, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol 2014;15:852-61. [Crossref] [PubMed]
- Valle J, Wasan H, Palmer DH, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med 2010;362:1273-81. [Crossref] [PubMed]
- Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434:913-7. [Crossref] [PubMed]
- Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917-21. [Crossref] [PubMed]
- de Bono J, Ramanathan RK, Mina L, et al. Phase I, Dose-Escalation, Two-Part Trial of the PARP Inhibitor Talazoparib in Patients with Advanced Germline BRCA1/2 Mutations and Selected Sporadic Cancers. Cancer Discov 2017;7:620-9. [Crossref] [PubMed]
- Cheng Y, Zhang J, Qin SK, et al. Treatment with olaparib monotherapy for BRCA2-mutated refractory intrahepatic cholangiocarcinoma: a case report. Onco Targets Ther 2018;11:5957-62. [Crossref] [PubMed]
- Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta 1998;1378:F115-77. [PubMed]
- Serrano M, Lee H, Chin L, et al. Role of the INK4a locus in tumor suppression and cell mortality. Cell 1996;85:27-37. [Crossref] [PubMed]
- Walker GJ, Gabrielli BG, Castellano M, et al. Functional reassessment of P16 variants using a transfection-based assay. Int J Cancer 1999;82:305-12. [Crossref] [PubMed]
- Zhang B, Peng Z. Defective folding of mutant p16(INK4) proteins encoded by tumor-derived alleles. J Biol Chem 1996;271:28734-7. [Crossref] [PubMed]
- Olivier M, Taniere P. Somatic mutations in cancer prognosis and prediction: lessons from TP53 and EGFR genes. Curr Opin Oncol 2011;23:88-92. [Crossref] [PubMed]
- Berge EO, Huun J, Lillehaug JR, et al. Functional characterisation of p53 mutants identified in breast cancers with suboptimal responses to anthracyclines or mitomycin. Biochim Biophys Acta 2013;1830:2790-7. [Crossref] [PubMed]
- Wagner PL, Stiedl AC, Wilbertz T, et al. Frequency and clinicopathologic correlates of KRAS amplification in non-small cell lung carcinoma. Lung Cancer 2011;74:118-23. [Crossref] [PubMed]
- Mita H, Toyota M, Aoki F, et al. A novel method, digital genome scanning detects KRAS gene amplification in gastric cancers: involvement of overexpressed wild-type KRAS in downstream signaling and cancer cell growth. BMC Cancer 2009;9:198. [Crossref] [PubMed]
- Pulciani S, Santos E, Long LK, et al. ras gene Amplification and malignant transformation. Mol Cell Biol 1985;5:2836-41. [PubMed]
(English Language Editor: R. Scott)



