Propofol protects hippocampal neurons in sleep-deprived rats by inhibiting mitophagy and autophagy
Introduction
Sleep deprivation (SD) can lead to disturbances in the energy metabolism of nerve cells (1,2), and intermittent sleep causes changes in energy metabolism in the brain. Chronic SD is known to cause mild neuroinflammation in rats, which in turn shows anxiety, decreased learning and memory (3). Even short-term SD has been found to impair the plasticity and memory of hippocampal neurons (4). SD can cause mitochondrial damage and oxidative stress in the hippocampus of rats (5), has been shown to reduce the membrane excitability of CA1 pyramidal neurons, and can induce Bax to migrate from the cytoplasm to the mitochondria, resulting in the release of cytochrome C into the cytoplasm (6).
Autophagy is a highly conservative biological process in which cells capture and degrade their cytoplasmic components. Under physiological conditions, the main purpose of autophagy activation is to adjust intracytoplasmic components and to regulate energy metabolism and maintain cellular homeostasis to promote cell survival (7). Recently, studies have reported that sleep deprivation may cause autophagy disorders in hippocampal neurons (8). An imbalance of autophagy may also affect long-term spatial memory formation in male rats (9). However, autophagy is regulated by a range of different factors (10), and its insufficient and excessive levels are harmful to neurons in the hippocampus (11). Mitophagy is a form of selective macro-autophagy resulting in damaged mitochondria being surrounded by autophagosomes. While mitochondrial damage induces apoptosis, milder mitochondrial damage can also induce autophagy (12). During stress, reactive oxygen species (ROS) are released from both the endoplasmic reticulum and mitochondria and can cause damage to the mitochondrial membrane (13). When cellular metabolism is abnormal or under environmental stress, mitochondria will fissure into populations of both damaged and healthy types. While healthy mitochondria fuse with other healthy mitochondria in a process that is mediated by mitochondrial fusion proteins, damaged mitochondria trigger the process of mitophagy and are removed from the cell (14). In brief, the mitochondrial quality control system maintains the dynamic balance of mitochondria via fusion and fission, which depends on the metabolic needs of cells under various physiological and pathological conditions.
PTEN-induced putative kinase 1 (PINK1) is a serine/threonine-protein kinase induced by PTEN capable of removing damaged mitochondria by inducing mitophagy to maintain mitochondrial stability. Parkin is a cytoplasmic E3 ligase, and its absence can cause abnormal mitochondrial morphology. While these two proteins are the key to mitophagy, the effect of SD on mitophagy has not yet been reported.
Propofol is widely used as an intravenous anesthetic, and with a molecular structure similar to phenolic antioxidants, can also exert an antioxidant effect (15). Natural sleep, in common with states of anesthesia, is actively generated by the brain, suggesting a potential overlap in neuronal circuitry (16). By analyzing the high-density EEG performance of both propofol-induced anesthesia and natural sleep, researchers have forwarded the hypothesis that propofol-induced general anesthesia resembles a sleep-like state (17). Propofol has been used to treat patients with refractory chronic primary insomnia, and found to be safe and effective, and improved the sleep quality of patients for a long period after treatment (18). The sleep produced by propofol not only causes effects associated with SD (19) but can also rescue the sleep debt of rats following SD (20). In a previous study, Cui and colleagues subjected PC12 nerve cells to oxygen deprivation and found that the level of autophagy was significantly increased. However, propofol intervention was shown to induce a reduction in the expression of autophagy-related proteins concomitant with an increase in nerve cell survival, indicating it had a protective effect on nerves (21). However, the neuroprotective effect of propofol in SD remains unclear.
In this study we aimed to understand the specific effect of propofol-induced sleep on the hippocampus of rats after SD and identify the potential mechanisms underlying this effect.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3872).
Methods
Animals and treatment
Male Sprague-Dawley rats were obtained from the Animal Experimental Center of Guangxi Medical University, China (weight: 260–320 g; age: 9–12 weeks-of-age) and were maintained in its Animal Experiment Center. All animal procedures were approved by the Animal Ethics Committee of Guangxi Medical University (No. 2017050029, Nanning, Guangxi, China), in compliance with the animal experiment specifications of Guangxi Medical University and the experimental animal welfare ethical review guidelines (GB/T 35892-2018) for the care and use of animals. A protocol was prepared before the study without registration.
To avoid interference between experimental and control groups, rats in the normal control group were kept in a separate room, while conditions in the two experimental animal rooms were standardized concerning sound, light, temperature, and humidity. Lights in the rooms (provided by a 40W incandescent lamp) were automatically turned on from 6:30 a.m. to 6:30 p.m., then turned off automatically from 6:30 p.m. to 6:30 a.m., and the ambient temperature was set to 23±2 °C and humidity 55%±15%. Each rat was labeled and numbered with an odor-free color on its back.
To evaluate the effect of different doses of propofol on the expression of Beclin1 protein in SD rats, we randomly them into a control group, SD group, propofol (5 mg/kg) group, propofol (10 mg/kg) group, propofol (15 mg/kg) group, propofol (30 mg/kg) group, and a propofol (60 mg/kg) group. Three rats were allocated to each of the eight groups. Rats in the control group (without SD) were fed by conventional methods, while those in all other groups used the modified multi-platform SD method for SD for a total period of 4 days (96 hours). Following the completion of SD, rats were dosed and subsequently tested. Rats in the control and SD group were given 1 mL of normal saline via intraperitoneal injection, while those in the propofol groups were given intraperitoneal injections of propofol at different doses (5, 10, 15, 30, and 60 mg/kg) after SD.
To confirm the effect of the autophagy pathway on propofol in reducing hippocampal damage, we randomly divided 112 rats into a control group (C group), SD group, SD + intralipid group (SD-F group), SD + propofol (30 mg/kg) group (P group), SD + rapamycin (0.5 mg/kg) group (R group), SD + rapamycin (0.5 mg/kg) + propofol (30 mg/kg) group (PR group), SD + 3-methyladenine (3-MA) (0.5mg/kg) group (3-MA group), and SD + 3-MA (0.5 mg/kg) + propofol (30 mg/kg) group (P + 3-MA group). Fourteen rats were allocated to each of the eight groups. Following SD, rats were given intraperitoneal injections of 1 mL of normal saline, 1 mL of 10% intralipid, 30 mg/kg of propofol, 0.5 mg/kg of rapamycin, and 0.5 mg/kg of 3-MA. Rats in the SD + rapamycin (0.5 mg/kg) + propofol (30 mg/kg) group and SD + 3-MA (0.5 mg/kg) + propofol (30 mg/kg) group were given intraperitoneal injections of 30 mg/kg of propofol half an hour after the injection of rapamycin or 3-MA.
To investigate the role of the PINK1/Parkin mitophagy pathway in the restorative effect of propofol on hippocampal mitochondrial damage after SD, 45 rats were randomly divided a control group, SD group, propofol (5 mg/kg) group, propofol (30 mg/kg) group, and propofol (60 mg/kg) group, with nine rats in each of the five groups. After SD, rats were given intraperitoneal injections of 1 mL of normal saline, 1 mL of 10% intralipid, 30 mg/kg of propofol, or 60 mg/kg of propofol.
Establishment of a rat model of rapid eye movement (REM) sleep deprivation
We used the modified multi-platform SD method to establish a rat model of rapid eye movement (REM) SD (22,23). The core mechanism underlying this technique is that body muscle tension will reduce after the animals enter the REM sleep period. The water level of the device was set in a suitable position so that the rats would still need to maintain a certain amount of neck muscle tension to avoid their nose touching the water when awake. If they entered the REM sleep period due to a reduction in muscle tension, they developed head ptosis, causing their nose to touch the water, waking the animal up, resulting in a REM SD effect.
The device was made of plexiglass (5 mm thick), measured 95 cm × 75 cm × 60 cm, and contained 12 small platforms with a diameter of 5 cm, height of 15 cm, and platform interval of 15 cm. Rats could move freely between each platform and the platform surface. The water level was 2 cm higher than the platform, forcing the rats to stand on the platform while maintaining a head-up position to avoid the nostrils falling into the water. The temperature of the water in the device was maintained at 23±1 °C with a thermostat. Sterile drinking water and food were provided at the top of the device and were available ad libitum.
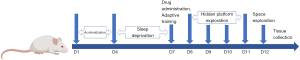
The water in the platform of the device was replaced at regular times (12 noon to 1 PM) and the device was cleaned carefully, during which time all rats were moved to a clean and dry box. While rats could move freely, eat, and drink, they were not permitted to sleep. If rats were found to be still and closed their eyes for more than 1 minute, the box was shaken until they were awoken. After cleaning, rats were moved back to the platform. The control group was placed in the same device, but without water. Rats in the control group were also allowed to sleep. Prior to the SD experiment, all rats were acclimatized to the device environment for 15 minutes, once a day for 3 days. The timeline of sleep deprivation, drug intervention, and behavioral experiments is shown in Figure 1.
Morris water maze
The Morris water maze device (Institute of Materials, Chinese Academy of Medical Sciences) consists of a water tank, an automatic track imaging recording device (with a camera fixed on the top of the water tank), and a computer with a track analysis and statistical analysis system (24,25). A round barrel stainless steel tank with a diameter of 1.5 m, a height of 45 cm, and a water depth of 20 cm was also used. The tank was divided into four quadrants, with each quadrant of the inner wall marked with four different shapes using black tape, and the water temperature was maintained at 23–25 °C. A platform was fixed in the southwest quadrant and submerged 2 cm below the surface, and the position of the platform remained unchanged for five consecutive days. A small amount of black ink was injected into the water to prevent rats from seeing the platform directly. During the experiment, a yellow shading cloth was used to surround the water tank to prevent the effect of automatic track recording from being disturbed by light and shadows arising from waves in the water.
Hidden platform exploration tests
During training, the rats were placed, in turn, into the water from four different quadrants (southwest → northwest → northeast → southeast) and were allowed to locate the submerged platform in the southwest quadrant of the tank. If a rat failed to find the submerged platform within 120 seconds, an assistant guided it towards the platform. After standing on the platform for 10 seconds, the rats were moved to a safe, dry, and warm place, to allow rest for two minutes before the next session. Escape latency was defined as the time for rats to find the submerged platform four times a day and was averaged over the four replicates.
Space exploration tests
This experiment was conducted on the 6th day after SD. After the submerged platform in the southwest quadrant was removed, the rats were placed into the water from the northwest quadrant. The swimming track of the rats was then recorded for over 60 seconds. Then recorded the retention time in the southwest quadrant and the number of times in which the rats crossed the position of the removed platform. This experiment was only performed once for each rat.
Transmission electron microscopy
All rats were sacrificed immediately after the intervention treatment, with the hippocampus removed then soaked in 3% glutaraldehyde at 4 °C for 2 hours. Hippocampal tissue was then cut into inducing 2 mm × 2 mm ×2 mm pieces then dehydrated step by step using ethanol and acetone. Finally, the specimens were embedded in epoxy resin, stained with uranyl acetate and lead citrate, and observed using an H‐7650 electron microscope (Hitachi, Tokyo, Japan).
Immunohistochemistry
Brain tissue was embedded in wax blocks, cut into 4-µm sections (Leica, China), and stored in a refrigerator at 4 °C. When preparing the experiment, the sections were warmed to room temperature and dewaxed to water, then incubated in 3% hydrogen peroxide for 20 minutes and transferred to a tris buffered saline (TBS)-based blocking solution containing 5% bovine serum albumin and 0.1% Tween-20 for 12 minutes. Sections were then incubated with a rabbit LC3A/B monoclonal antibody (1:500, Cell Signaling Technology, USA), a rabbit Pink1 polyclonal antibody (1:100, Affinity Biosciences. the USA), and a rabbit PARK2/Parkin polyclonal antibody (1:200, Proteintech Group, USA) at 4 °C overnight. The following morning, sections were washed with PBS then incubated in IRDye 680RD Goat anti-Rabbit IgG (1:500, LI-COR Biosciences, USA) at room temperature for 12 minutes. Sections were then treated with Streptavidin/HRP and incubated for 10 minutes. Finally, the sections were washed with PBS, developed with 3,3-diaminobenzidine (DAB), stained with Hematoxylin, and microscopically examined. Fluorescent images were taken with an Olympus VS120 Virtual Slide Scanner.
Western blotting
Shredded hippocampus tissue was homogenized in a lysis buffer containing RIPA lysis buffer (Roche, USA), phenylmethylsulfonyl fluoride (PMSF), and phosphatase inhibitors (Solarbio, Beijing, China) in a one-to-one ratio. After 30 minutes of lysis, the samples were centrifuged at 12,000 rpm for 5min at 4 °C, and the supernatants removed. A protein standard curve was then prepared using a bovine serum albumin standard solution (1 mg/mL), and the protein concentration of the supernatants was determined by a microplate reader using a bovine serum protein concentration determination kit (Solarbio, Beijing, China). Proteins were then separated using 12% SDS-PAGE, transferred to a PVDF membrane, then incubated overnight at 4 °C with primary antibodies, including rabbit LC3A/B monoclonal antibody (1:1,000, Cell Signaling Technology, USA), rabbit Pink1 polyclonal antibody (1:1,000, Affinity Biosciences, USA), rabbit PARK2/Parkin polyclonal antibody (1:1,000, Proteintech Group, USA), rabbit Beclin1 monoclonal antibody (1:1,000, Cell Signaling Technology, USA), and rabbit p62 polyclonal antibody (1:1,000, Cell Signaling Technology, USA). IRDye 680RD Goat anti-Rabbit IgG (1:15,000, LI-COR Biosciences, USA) was then added and incubated at room temperature for 2 hours. The PVDF membranes were then scanned with the Odyssey two-color fluorescence imaging system and the bands were analyzed using Odyssey (LI-COR Biosciences, USA) and Image J software (NIH, USA). The ratio of the gray value of the target protein band to the gray value of the internal reference (GAPDH) band was expressed as the relative expression level of the protein.
RT-PCR
Total RNA was isolated using a Total RNA Extraction Kit 9108-9109 (Takarabiomed, Beijing, China), and a reverse transcription Kit (RR047A, Takarabiomed, Beijing, China) was used for cDNA synthesis. Quantitative RT-PCR for the genes of interest was then performed with an Amplification kit (RR820A, Takarabiomed, Beijing, China) and a PCR machine (Thermo Scientific, USA) following the manufacturer’s protocols. Primers were provided by Sango Biotech (Shanghai, China), and the primer pairs for each gene of interest were as follows: PINK1: forward 5'-CATGGCTTTGGATGGAGAGT-3', reverse 5'-TGGGAGTTTGCTCTTCAAGG-3'; Parkin: forward 5'-CTGGCAGTCATTCTGGACAC-3', reverse 5'-CTCTCCACTCATCCGGTTTG-3'; LC3: forward 5'-CTTCGCCGACCGCTGTAA-3', reverse 5'-ATCCGTCTTCATCCTTCTCCTG-3'; GAPDH: forward 5'-ACAGCAACAGGGTGGTGGAC-3', reverse 5'-TTTGAGGGTGCAGCGAACTT-3'.
Statistical analyses
All statistical analyses were performed with SPSS version 22.0 (IBM, USA) and GraphPad Prism version 8.3 (GraphPad Software, USA) software. All measurement data are expressed as mean ± standard deviation (SD) and were tested for normal distributions and homogeneity of variance. The results of the Morris water maze behavior experiment were tested by multiple factor analysis of variance (ANOVA) for repeated measurements, and one-way ANOVA was used for intra-group comparisons. The least significant difference (LSD) test was used for comparisons between groups and P<0.05 was considered to represent statistical significance.
Results
Effect of different doses of propofol on Beclin1 protein expression in SD rats
Following SD, rats in each group received an intraperitoneal injection of propofol at a concentration of 5, 10, 15, 30, 45, or 60 mg/kg. The expression of Beclin1 protein was then detected after the rats were awoken.
As shown in Figure 2, the expression of Beclin1 protein in rats was significantly increased after SD. However, when propofol administration reached a concentration of 30 mg/kg in SD rats, a significant reduction in the expression levels of Beclin1 protein was observed, although, when the dose of propofol was increased, the trend for reduction did not increase linearly. This showed 30 mg/kg was the lowest effective dose of propofol that can reduce autophagic damage in SD rats and we used this dose as an interventional factor to influence autophagy in the next experiment.

Role of the autophagy pathway in the propofol-induced alleviation of hippocampal injury in rats following SD
To further determine the mechanism by which propofol reduced autophagy caused by SD, we used the autophagy enhancer rapamycin, and the autophagy inhibitor 3-methyladenine (3-MA), as interventional factors in a rat model of SD.
Rats were injected intraperitoneally with rapamycin or 3-MA after SD, then with or without 30 mg/kg propofol 30 minutes later. Various tests and inspections were conducted 1 hour after the rats had been awakened, and as shown in Figure 3, the expression levels of Beclin1 and LC3 in the group which were administered propofol or/and the 3-MA were significantly lower than those in the SD group and the R group. However, propofol did not reverse the effect of rapamycin in the PR group and did not enhance the effect of 3-MA in the P + 3-MA group.

We also used transmission electron microscopy to observe (×25,000) mitochondria in the hippocampal neurons, as shown in Figure 4. This showed that after SD, the hippocampal membrane structure was surrounded by phagocytic bubbles, some of which were typical peanut-shaped, the mitochondrial crest shape in the mitochondria was blurred, and in some cases, mitochondrial fragmentation was evident. SD rats injected with propofol and/or 3-MA showed significant improvement with regards to this phenomenon, while those injected with propofol and/or rapamycin did not show any significant improvement.

Behavioral experiments carried out with the Morris water maze showed similar results to those listed above (Figure 5). The results of the fifth day of the hidden platform test showed that compared with the C group, the escape latency of rats in the SD group, SD-F group, PR group, and R group was significantly longer, while there was no significant difference in those in group P, P + 3-MA, and 3-MA group compared with the C group. Rats in the P group, 3-MA group, and P + 3-MA group displayed shorter escape latency compared with the SD group on the fifth day of the hidden platform test (Figure 5B), while those in the SD group, SD-F group, PR group, and R group crossed the hidden platform fewer times and stayed in the target quadrant for a shorter time in contrast with the control group (Figure 5C). However, propofol or/and 3-MA administration alleviated the impaired memory of SD rats significantly. As shown in Figure 5C, rats in the P group and P + 3-MA group passed through the hidden platform more times and stayed in the target quadrant for a longer time, compared with the SD group. The representative swimming tracks of rats in each group are illustrated in Figure 5D to show the difference among the groups. Collectively, these data suggested SD aggravated cognitive impairment in rats, while propofol and 3-MA alleviated it.

Accordingly, these results suggested several conclusions. First, that SD could lead to enhanced autophagy; second, that propofol and 3-MA could reduce autophagy and mitochondrial damage in hippocampal neurons caused by SD, which was an effect similar to that of 3-MA; and third, the effect of propofol could be reversed by rapamycin.
Role of the mitophagy pathway in the propofol-induced alleviation of hippocampal mitochondrial damage in rats after SD
The above transmission electron microscopy results (Figure 4) suggest SD caused morphological damage in the hippocampal mitochondria of rats after SD and that propofol reduced this damage.
We next hypothesized that propofol alleviated hippocampal mitochondrial damage after SD in rats via the PINK1/Parkin mitophagy signaling pathway. As shown in Figure 6A-6C, LC3, PINK1, and Parkin expression levels were detected using qRT-PCR, and the results demonstrate the expressions of mRNA of LC3, PINK1, and Parkin in the group receiving propofol were significantly lower than those in the SD group not receiving it. Furthermore, the expression of proteins of LC3, PINK1, and Parkin in the hippocampus measured by immunohistochemistry produced similar results (Figure 6D-6I). Moreover, the administration of propofol could reduce the expression levels of PINK1, P62, LC3, and Beclin1 protein, measured by Western blotting, after SD (Figure 6J-6O).

In summary, our data indicated SD also caused an increase in excessive mitophagy and autophagy-related proteins, although this experiment did not show a dose-dependent effect on propofol.
Discussion
Autophagy is an important mechanism for maintaining cellular homeostasis. Dysregulated autophagy may be involved in many pathophysiological processes (8,26), and sleep fragmentation can also change the rhythm of hippocampal autophagy (27). Our data show that 96 hours of SD could simultaneously lead to the increased expression of autophagy-related proteins (Beclin1, LC3, and p62) in hippocampal cells, and impair learning, cognition, and memory in rats (Figures 2,5,6). Homologous to the yeast Atg6, Beclin1is a key protein that mediates the localization of other autophagy proteins to pre-autophagosomes and combines with vacuolar protein sorting 34 (Vsp34) to form a complex that induces autophagy. Thus, Beclin1 is an important regulator of mammalian autophagosome formation (28). LC3 (microtubule-associated protein light chain 3, MAP LC3) was the first autophagosome marker protein to be identified in mammals and is homologous to the yeast autophagy-related gene Atg8. Similar to previous studies, SD has been shown to cause excessive levels of autophagy and may be one of the causes of impairment of learning and memory (8,29,30).
Our study also observed obvious autophagic vesicles in rat hippocampal mitochondria after SD, then detected the relative expression of several mitophagy-related proteins (PINK1, Parkin, and p62), which are the initiating proteins of mitophagy (Figures 4,6). This suggests SD induces excessive mitophagy at the same time as autophagy occurs, and both can be caused by mitochondrial dysfunction. However, previous studies have not reported the effect of SD on mitophagy. SD or sleep disorders can alter energy metabolism in the brain and affect glycogen transport and uptake (2,3,6), and it is well known that SD can cause an increase in ROS and damage to mitochondria, which in turn induces apoptosis (5,31,32). Compared with other organelles, mitochondria often change shape under physiological and pathological conditions. However, once mitochondria are damaged by stress levels which are beyond their ability to withstand, a greater response is activated in terms of degradation, that is, mitophagy. The mitochondrial quality control system can repair partially damaged mitochondria and clear those which cannot be repaired. PINK1, located in the outer membrane of the mitochondria, accumulates and recruits Parkin during mitochondrial injury, which can cause ubiquitination in the mitochondrial stroma and ultimately remove damaged mitochondria (33,34). Parkin on the mitochondrial membrane is activated by phosphorylation by PINK1, which in turn induces the ubiquitination and aggregation of E3, causing ubiquitinated mitochondria to be recognized by connexin p62. As a multifunctional adaptor protein, p62/SQSTM1 mediates the binding of ubiquitin to autophagosomes, and the combination of p62 and LC3 connects mitochondria to the autophagic vesicle membrane and causes mitophagy (34-36). Both in vivo and in vitro models of stroke, excessive activation of mitophagy will lead to increased neuronal death (37,38). In another animal experiment, excessive mitophagy aggravated the brain damage in rats following cardiac arrest and cardiopulmonary resuscitation (39). Therefore, excessive mitophagy may be related mitochondrial damage and dysfunction by excessive levels of stress caused by SD and may also be one of the causes of learning and memory impairment.
As an antioxidant, propofol can be activated by inhibiting NADPH oxidase-mediated mast cell degranulation, protecting intestinal epithelial cells from ischemia-reperfusion injury. It can also inhibit the expression of the autophagy-related proteins LC3 and Beclin1 induced by lipopolysaccharides (40). Studies have shown that propofol can inhibit the overexpression of synaptophysin by inhibiting excessive autophagy, protecting the learning and memory abilities of depressed rats after Electroconvulsive shock (41). Similar to previous research, the results of the present study showed propofol intervention could reduce the expression levels of autophagy-related genes and proteins after SD in rats and improve the morphology of the mitochondria to a normal state. It has been suggested that propofol intervention could alleviate the autophagy of the rat hippocampus caused by SD. At the same time, after the administration of propofol, the learning and memory impairment caused by SD was improved.
Mammalian target of rapamycin (mTOR) is involved in neural development, synaptic plasticity, learning, and memory, and can combine with other proteins to form two complexes with different functions: mTORC1 and mTORC2. Activated mTORC1 phosphorylates serine/threonine kinase UNC51-like kinase 1 and autophagy-related protein 13, preventing it from forming a complex with the pre-autophagosomal structure, thereby inhibiting autophagy. Based on this, it was considered to be the core protein controlling autophagy (42). Rapamycin is currently the most widely used autophagy enhancer (43). It inhibits the activity of mTORC1 by binding to the FKBP12 region on mTORC1, thereby activating autophagy. 3-MA is a class III phosphoinositide 3-kinase inhibitor and predominantly acts via the activity of Beclin1 and the Vps34 complex to inhibit autophagy (44).
To further prove that its effect on improving learning and memory impairment was by inhibiting excessive autophagy, we use rapamycin and 3-MA to block or replace the effect of propofol, and the results suggested its effect could be reversed by rapamycin in rats after SD. This suggests the impairment of hippocampal function in rats after SD may be related to the excessive activation of autophagy. Rapamycin reversed the effect of propofol that inhibited the autophagy of hippocampal cells during anesthesia in elderly rats and 6-day-old rats (45,46). On the other hand, in our experiments, 3-MA reduced autophagy and improved behavioral experimental results. This suggested the improvement of function in the rat hippocampus after SD may be related to the inhibition of autophagy (44). The experimental results of Cui et al. also suggest that both propofol and 3-MA significantly inhibited the expression of autophagy-related proteins in hippocampal cells caused by ischemia-reperfusion (47). Propofol has also been shown to alleviate autophagy and apoptosis induced by ischemia-reperfusion by inhibiting the expression of autophagy-related proteins such as Beclin1, LC3, and mTOR (48). These findings indicated propofol prevented memory and learning deficits induced by SD, by inhibiting excessive autophagy.
In our experiments, the mRNA and protein levels of PINK1 and Parkin in the hippocampus of rats increased after SD and decreased after propofol intervention. This suggests that propofol may play a role through the PINK1/Parkin mitophagy pathway. Propofol inhibits autophagy while inhibiting excessive mitophagy to maintain the balance of cellular mitochondrial metabolism. These effects may be another reason why propofol alleviates learning and memory impairment caused by SD. Propofol has been reported to participate in the regulation of autophagy and the protection of mitochondria. Its pretreatment can also inhibit the expression of endothelial microvesicles and microcystin-1 induced by hypoxia/reoxygenation in human umbilical vein endothelial cells. Moreover, propofol pretreatment also reduced mitochondrial damage caused by stress by inhibiting the mitochondrial apoptosis pathway (49), and in a previous study, Liang (50) found that propofol reduced the expression of PINK1 in the hippocampal cells of newborn rats.
However, it is important to note that autophagy is a common life phenomenon for eukaryotes, and its existence is limited to more or less rather than existence or absence. Excessive restrictions on autophagy or enhanced autophagy may also cause new problems.
In the present study, we confirmed that propofol inhibited the increased expression of autophagy-related and mitophagy-related proteins in the rat hippocampus after SD, thereby inhibiting autophagy and mitophagy in hippocampus cells. These results indicate that SD can lead to enhanced autophagy and mitophagy in the rat hippocampus and may provide an experimental basis for propofol to be incorporated into new clinical applications.
Acknowledgments
We are grateful to the staff involved in this research, for their concern, support, and understanding during our experiment and writing.
Funding: This work was supported by the Key Research and Development program of Guangxi (No. AB18221031), Natural Science Foundation of Guangxi (No. 2020GXNSFDA238025), and National Natural Science Foundation of China (No. 81373498).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3872
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-3872
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3872). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal procedures were approved by the Animal Ethics Committee of Guangxi Medical University (No. 2017050029, Nanning, Guangxi, PR China), in compliance with the animal experiment specifications of Guangxi Medical University and the experimental animal welfare ethical review guidelines (GB/T 35892-2018) for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hipólide DC, Suchecki D, Pimentel de Carvalho Pinto A, et al. Paradoxical sleep deprivation and sleep recovery: effects on the hypothalamic-pituitary-adrenal axis activity, energy balance and body composition of rats. J Neuroendocrinol 2006;18:231-8. [Crossref] [PubMed]
- Xu X, Wang R, Sun Z, et al. Trehalose enhances bone fracture healing in a rat sleep deprivation model. Ann Transl Med 2019;7:297. [Crossref] [PubMed]
- Manchanda S, Singh H, Kaur T, et al. Low-grade neuroinflammation due to chronic sleep deprivation results in anxiety and learning and memory impairments. Mol Cell Biochem 2018;449:63-72. [Crossref] [PubMed]
- Prince TM, Wimmer M, Choi J, et al. Sleep deprivation during a specific 3-hour time window post-training impairs hippocampal synaptic plasticity and memory. Neurobiol Learn Mem 2014;109:122-30. [Crossref] [PubMed]
- Salehpour F, Farajdokht F, Erfani M, et al. Transcranial near-infrared photobiomodulation attenuates memory impairment and hippocampal oxidative stress in sleep-deprived mice. Brain Res 2018;1682:36-43. [Crossref] [PubMed]
- Yang RH, Hu SJ, Wang Y, et al. Paradoxical sleep deprivation impairs spatial learning and affects membrane excitability and mitochondrial protein in the hippocampus. Brain Res 2008;1230:224-32. [Crossref] [PubMed]
- Menzies FM, Fleming A, Caricasole A, et al. Autophagy and Neurodegeneration: Pathogenic Mechanisms and Therapeutic Opportunities. Neuron 2017;93:1015-34. [Crossref] [PubMed]
- Cao Y, Li Q, Liu L, et al. Modafinil protects hippocampal neurons by suppressing excessive autophagy and apoptosis in mice with sleep deprivation. Br J Pharmacol 2019;176:1282-97. [Crossref] [PubMed]
- Hylin MJ, Zhao J, Tangavelou K, et al. A role for autophagy in long-term spatial memory formation in male rodents. J Neurosci Res 2018;96:416-26. [Crossref] [PubMed]
- Yamamoto A, Yue Z. Autophagy and its normal and pathogenic states in the brain. Annu Rev Neurosci 2014;37:55-78. [Crossref] [PubMed]
- Li L, Zhang Q, Tan J, et al. Autophagy and hippocampal neuronal injury. Sleep Breath 2014;18:243-9. [Crossref] [PubMed]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol 2011;12:9-14. [Crossref] [PubMed]
- Xiao B, Goh JY, Xiao L, et al. Reactive oxygen species trigger Parkin/PINK1 pathway-dependent mitophagy by inducing mitochondrial recruitment of Parkin. J Biol Chem 2017;292:16697-708. [Crossref] [PubMed]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science 2012;337:1062-5. [Crossref] [PubMed]
- Li W, Zhang Y, Liu Y, et al. In vitro kinetic evaluation of the free radical scavenging ability of propofol. Anesthesiology 2012;116:1258-66. [Crossref] [PubMed]
- Lydic R, Biebuyck JF. Sleep neurobiology: relevance for mechanistic studies of anaesthesia. Br J Anaesth 1994;72:506-8. [Crossref] [PubMed]
- Murphy M, Bruno MA, Riedner BA, et al. Propofol anesthesia and sleep: a high-density EEG study. Sleep 2011;34:283-91A. [Crossref] [PubMed]
- Xu Z, Jiang X, Li W, et al. Propofol-induced sleep: efficacy and safety in patients with refractory chronic primary insomnia. Cell Biochem Biophys 2011;60:161-6. [Crossref] [PubMed]
- Tung A, Lynch JP, Mendelson WB. Prolonged sedation with propofol in the rat does not result in sleep deprivation. Anesth Analg 2001;92:1232-6. [Crossref] [PubMed]
- Tung A, Bergmann BM, Herrera S, et al. Recovery from sleep deprivation occurs during propofol anesthesia. Anesthesiology 2004;100:1419-26. [Crossref] [PubMed]
- Cui D, Wang L, Qi A, et al. Propofol prevents autophagic cell death following oxygen and glucose deprivation in PC12 cells and cerebral ischemia-reperfusion injury in rats. PLoS One 2012;7:e35324 [Crossref] [PubMed]
- van Hulzen ZJ, Coenen AM. Paradoxical sleep deprivation and locomotor activity in rats. Physiol Behav 1981;27:741-4. [Crossref] [PubMed]
- Machado RB, Hipólide DC, Benedito-Silva AA, et al. Sleep deprivation induced by the modified multiple platform technique: quantification of sleep loss and recovery. Brain Res 2004;1004:45-51. [Crossref] [PubMed]
- Morris RG, Garrud P, Rawlins JN, et al. Place navigation impaired in rats with hippocampal lesions. Nature 1982;297:681-3. [Crossref] [PubMed]
- Vorhees CV, Williams MT. Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat Protoc 2006;1:848-58. [Crossref] [PubMed]
- Yu L, Chen Y, Tooze SA. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2018;14:207-15. [Crossref] [PubMed]
- He Y, Cornelissen-Guillaume GG, He J, et al. Circadian rhythm of autophagy proteins in hippocampus is blunted by sleep fragmentation. Chronobiol Int 2016;33:553-60. [Crossref] [PubMed]
- Di Rita A, Peschiaroli A, D, Acunzo P, et al. HUWE1 E3 ligase promotes PINK1/PARKIN-independent mitophagy by regulating AMBRA1 activation via IKKα. Nat Commun 2018;9:3755. [Crossref] [PubMed]
- Yang SQ, Jiang L, Lan F, et al. Inhibited Endogenous H2S Generation and Excessive Autophagy in Hippocampus Contribute to Sleep Deprivation-Induced Cognitive Impairment. Front Psychol 2019;10:53. [Crossref] [PubMed]
- Li Y, Zhang Y, Ji G, et al. Autophagy Triggered by Oxidative Stress Appears to Be Mediated by the AKT/mTOR Signaling Pathway in the Liver of Sleep-Deprived Rats. Oxid Med Cell Longev 2020;2020:6181630 [Crossref] [PubMed]
- Chong SJ, Low IC, Pervaiz S. Mitochondrial ROS and involvement of Bcl-2 as a mitochondrial ROS regulator. Mitochondrion. 2014 Nov;19 Pt A:39-48.
- Somarajan BI, Khanday MA, Mallick BN. Rapid Eye Movement Sleep Deprivation Induces Neuronal Apoptosis by Noradrenaline Acting on Alpha1 Adrenoceptor and by Triggering Mitochondrial Intrinsic Pathway. Front Neurol 2016;7:25. [Crossref] [PubMed]
- Matheoud D, Sugiura A, Bellemare-Pelletier A, et al. Parkinson's Disease-Related Proteins PINK1 and Parkin Repress Mitochondrial Antigen Presentation. Cell 2016;166:314-27. [Crossref] [PubMed]
- Gladkova C, Maslen SL, Skehel JM, et al. Mechanism of parkin activation by PINK1. Nature 2018;559:410-4. [Crossref] [PubMed]
- Yoo SM, Jung YK. A Molecular Approach to Mitophagy and Mitochondrial Dynamics. Mol Cells 2018;41:18-26. [PubMed]
- Durcan TM, Fon EA. The three 'P's of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev 2015;29:989-99. [Crossref] [PubMed]
- Shi RY, Zhu SH, Li V, et al. BNIP3 interacting with LC3 triggers excessive mitophagy in delayed neuronal death in stroke. CNS Neurosci Ther 2014;20:1045-55. [Crossref] [PubMed]
- Deng Z, Ou H, Ren F, et al. LncRNA SNHG14 promotes OGD/R-induced neuron injury by inducing excessive mitophagy via miR-182-5p/BINP3 axis in HT22 mouse hippocampal neuronal cells. Biol Res 2020;53:38. [Crossref] [PubMed]
- Huang Y, Gao X, Zhou X, et al. Mitophagy in the Hippocampus Is Excessive Activated After Cardiac Arrest and Cardiopulmonary Resuscitation. Neurochem Res 2020;45:322-30. [Crossref] [PubMed]
- Gan X, Xing D, Su G, et al. Propofol Attenuates Small Intestinal Ischemia Reperfusion Injury through Inhibiting NADPH Oxidase Mediated Mast Cell Activation. Oxid Med Cell Longev 2015;2015:167014 [Crossref] [PubMed]
- Li P, Hao XC, Luo J, et al. Propofol Mitigates Learning and Memory Impairment After Electroconvulsive Shock in Depressed Rats by Inhibiting Autophagy in the Hippocampus. Med Sci Monit 2016;22:1702-8. [Crossref] [PubMed]
- Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017;168:960-76. [Crossref] [PubMed]
- Bové J, Martínez-Vicente M, Vila M. Fighting neurodegeneration with rapamycin: mechanistic insights. Nat Rev Neurosci 2011;12:437-52. [Crossref] [PubMed]
- Miller S, Tavshanjian B, Oleksy A, et al. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science 2010;327:1638-42. [Crossref] [PubMed]
- Yang N, Li Z, Han D, et al. Autophagy prevents hippocampal α-synuclein oligomerization and early cognitive dysfunction after anesthesia/surgery in aged rats. Aging (Albany NY) 2020;12:7262-81. [Crossref] [PubMed]
- Cho S, Jung YJ, Suh EC, et al. The recovery from transient cognitive dysfunction induced by propofol was associated with enhanced autophagic flux in normal healthy adult mice. Brain Res 2018;1700:99-108. [Crossref] [PubMed]
- Cui DR, Wang L, Jiang W, et al. Propofol prevents cerebral ischemia-triggered autophagy activation and cell death in the rat hippocampus through the NF-κB/p53 signaling pathway. Neuroscience 2013;246:117-32. [Crossref] [PubMed]
- Noh HS, Shin IW, Ha JH, et al. Propofol protects the autophagic cell death induced by the ischemia/reperfusion injury in rats. Mol Cells 2010;30:455-60. [Crossref] [PubMed]
- Deng F, Wang S, Cai S, et al. Inhibition of Caveolae Contributes to Propofol Preconditioning-Suppressed Microvesicles Release and Cell Injury by Hypoxia-Reoxygenation. Oxid Med Cell Longev 2017;2017:3542149 [Crossref] [PubMed]
- Liang C, Du F, Cang J, et al. Pink1 attenuates propofol-induced apoptosis and oxidative stress in developing neurons. J Anesth 2018;32:62-9. [Crossref] [PubMed]