
Effect of mesenchymal-epithelial transition amplification on immune microenvironment and efficacy of immune checkpoint inhibitors in patients with non-small cell lung cancer
Introduction
In recent years, immune checkpoint inhibitors (ICIs), represented by anti-programmed cell death receptor ligand 1 (anti-PD-L1), its ligand PD-L1, and anti-cytotoxic T-lymphocyte-associated protein 4 (anti-CTLA-4), have opened a new chapter for the treatment of advanced non-small cell lung cancer (NSCLC). A series of clinical trials confirmed the therapeutic effect of ICIs in NSCLC (1), and in advanced NSCLC, the response rate of anti-PD-1/PD-L1 monotherapy was 17–21%, although some patients responded very persistently. Therefore, the identification of suitable biomarkers to screen the dominant population for immunotherapy efficacy is particularly important (2).
Increasingly, studies have shown that inflammatory tumor immune microenvironment is associated with the effectiveness of immunotherapy (3-7). High expression of PDL1, infiltration of inflammatory cells such as CD8+ T lymphocytes and Expression of inflammatory cytokines are all regarded as the important element of inflammatory tumor immune microenvironment (8-10). Therefore, the gene which can remodel the tumor immune microenvironment to be inflammatory type, it could be the efficacy biomarker to predict the efficacy of immunotherapy.
To date, studies have suggested that specific gene mutations may be potential predictors of ICI treatment response (11-14). TP53, mesenchymal-epithelial transition (MET) and KRAS driver mutations in lung adenocarcinoma (LUAD) have been found can regulate immune cell infiltration and PD-L1 expression, both of which may represent potential predictors guiding ICI treatment (11). Similarly, a recent study found that recurrent mutations in TET1 (TET1-MT) were predictive of a relatively good durable clinical benefit from ICIs and an improvement in overall survival (OS) across multiple cancer types (12).
High-level MET amplification, MET exon 14 skipping alterations (MET∆14), or MET overexpression are the different type of MET gene alternation. MET amplification has been reported as a valuable marker for poor prognosis in advanced unresectable tumor such as colorectal cancer, breast carcinoma and gastric carcinomas but not in NSCLC patients (15,16). Studies have suggested that patients with MET mutations and MET amplification may be more responsive to immunotherapy than to targeted therapy (15,17,18), as MET mutation, amplification, or overexpression can upregulate coinhibitory molecule expression and downregulate costimulatory molecule expression (17). However, most of these studies were based on the cell molecular biology and immunohistochemistry, lacking the data of clinical immunotherapy especially for Chinese patients. Therefore, the correlation between MET amplification and the efficacy of NSCLC immunotherapy remains unclear.
In this study, we collected copy number alteration (CNA), somatic mutation, and clinical data of patients with NSCLC who were treated with ICIs to further assess the correlation between MET amplification and the efficacy of ICIs in patients with the disease. We found that in patients with MET-amplified NSCLC, ICI treatment was associated with longer progression-free survival (PFS) times, enhanced immunogenicity, and activated antitumor immunity. Our study suggests that MET amplification may be a novel predictive marker for immunotherapy efficacy in NSCLC. We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/atm-21-4543).
Methods
Clinical cohorts and cancer cell lines
To assess the correlation between MET amplification and the efficacy of NSCLC immunotherapy, data from ICI-treated patients in two NSCLC clinical cohorts were collected and processed as shown in Figure S1. The first ICI-treated cohort from Rizvi et al. consisted mainly of 240 NSCLC samples (n=240) with CNA, somatic mutation, and clinical data (19). Samples from the Rizvi et al. cohort (n=240) were sequenced using the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) panel. As the ICI-treated cohort from Samstein et al. (20) consisted of only four patients with MET amplification, we collected CNA, tumor mutation burden (TMB), and clinical data from another ICI-treated cohort from Zhujiang Hospital of Southern Medical University, Guangzhou Chest Hospital and First People’s Hospital of Chenzhou. A total of 10 patients received ICIs (anti-PD-1 monotherapy, ≥3 treatment lines) to investigate the effect of MET amplification on the prognosis of NSCLC immunotherapy. Detailed clinical characteristics of patients with MET amplification are listed in https://cdn.amegroups.cn/static/public/atm-21-4543-1.docx. This research was performed in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the ethics committee of Zhujiang Hospital of Southern Medical University, Guangzhou Chest Hospital and First People’s Hospital of Chenzhou. Written informed consent was obtained from the individuals for the publication of any potentially identifiable images or data included in this article.
Clinical and sample data (mRNA expression and somatic mutation data) from The Cancer Genome Atlas (TCGA)-LUAD and TCGA-lung squamous cell carcinoma (LUSC) datasets were downloaded from the Genomic Data Commons (https://portal.gdc.cancer.gov/) using the R package TCGAbiolinks (21). The TCGA-LUSC and TCGA-LUAD datasets were combined into the TCGA-NSCLC dataset for subsequent analysis. Gene expression and protein expression levels in the TCGA-NSCLC dataset were log2 normalized [fragments per kilobase of exon model per million mapped fragments (FPKM) +1] and were expressed as the normalized reverse-phase protein array (RPPA) values.
In addition, we used cBioPortal (22) (https://www.cbioportal.org) to download CNA data from the TCGA-LUAD and TCGA-LUSC datasets. The units of CNA were GISTIC 2.0 copy number.
We downloaded CNA and drug sensitivity data for 69 NSCLC cell lines from the Genomics of Drug Sensitivity in Cancer (GDSC) database (23), and the units of CNA and drug sensitivity data were GISTIC 2.0 copy number and in half-maximal inhibitory concentration (IC50) value.
Clinical samples and classification of lung cancer cell lines
Consistent with a previous study (22), for the TCGA-NSCLC cancer immunotherapy datasets (from Rizvi et al. and the local cohort), a GISTIC score of ≥2 was defined as MET amplification, and a score of <2 was defined as non-MET amplification. Similarly, in the CNA data for the GDSC-NSCLC cell lines, a GISTIC score of ≥2 was defined as MET amplification, and a score of <2 was defined as non-MET amplification.
mRNA expression profiling, RPPA analysis, and drug sensitivity analysis
Methods for tumor RNA extraction, mRNA library preparation, sequencing, quality control, and subsequent data processing to quantify gene expression in TCGA-NSCLC samples have been previously reported (24). MET protein expression in the TCGA-NSCLC dataset was based on the RPPA of the TCGA database (24). Drug sensitivity data for human NSCLC cell lines are available from GDSC (https://www.cancerrxgene.org/).
Immunological characteristics and correlation analysis of tumor immunogenicity
We used the CIBERSORT web portal (http://cibersort.stanford.edu/) (25) to analyze mRNA expression data from the TCGA-NSCLC dataset to estimate the abundance of 22 immune cell types in the TCGA-NSCLC dataset. The list of immune-related genes and the neoantigen data for the TCGA-NSCLC dataset were obtained from Thorsson et al. (26), and the expression levels of these genes were quantified as log2 (FPKM +1) values.
TMB refers to the total number of substitutions and insertion/deletion mutations per megabase in the exon-coding region of a tumor gene in the tumor cell genome (27). TMB was defined as nonsynonymous mutations in the TCGA-NSCLC and local cohorts. We used the somatic mutation data in the TCGA-NSCLC dataset as the raw mutation count and 38 Mb as the estimate of the exome size (28).
Functional and pathway enrichment analyses
For gene annotation enrichment analysis using the clusterProfiler R package, differences in Gene Ontology (GO) terms, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and Reactome pathways for which P<0.05 were considered significant. The gene set enrichment analysis (GSEA) gene set was obtained from the Broad Institute Molecular Signatures Database (MSigDB) (29).
Gene set related to DNA damage response and repair (DDR) pathway analysis
We used the DDR pathway gene set from the Broad Institute MSigDB database (29), which includes the following eight DDR-related pathways: “R-HSA-6783310_REACTOME_Fanconi_Anemia_Pathway”, “hsa03450_KEGG_Non_Homologous_End_Joining”, “R-HSA-73884_REACTOME_Base_Excision_Repair”, “hsa03430_KEGG_Mismatch_Repair”, “R-HSA-5696398_REACTOME_Nucleotide_Excision_Repair”, “R-HSA-5696398_REACTOME_Double_Strand_Break_Repair”, “GO:0003697_Single_Stranded_DNA_Binding”, and “hsa03440_KEGG_Homologous_Recombination”. Detailed gene sets related to DDR pathway analysis are listed in https://cdn.amegroups.cn/static/public/atm-21-4543-2.xlsx.
Statistical analysis
The Mann-Whitney test was used to compare TMB, neoantigen load (NAL), immune cell content, and immune gene mRNA and protein content between the MET-amplification and non-MET-amplification groups. Fisher’s exact test was used to compare correlations in smoking history, clinical stage, and sex between the groups, and the correlation between the MET amplification status and sex was compared using the chi-square test. PFS and OS in the MET-amplification and non-MET-amplification groups were analyzed by the Kaplan-Meier method with a log-rank test and by Cox proportional hazards regression analysis. P<0.05 was considered statistically significant, and all statistical tests were two-sided. R software (version 3.6) was used for statistical analysis, and the R package “ComplexHeatmap” (30) was employed to visualize the mutation and immune cell landscape.
Results
Mutation profile of the study population
We used cBioPortal to obtain a published study of NSCLC immunotherapy from Rizvi et al. comprising 240 patients receiving ICI treatment [anti-PD-(L)1 monotherapy or combination treatment with anti-CTLA-4 therapy]. This included CNA, somatic mutation, and clinical data for patients with NSCLC, and was used to further investigate the critical role of MET amplification in the prognosis of NSCLC immunotherapy. In order to explored Chinese patients data, we integrated the copy number variation, somatic mutation, and clinical data of NSCLC patients (n=10) with MET amplification from Zhujiang Hospital of Southern Medical University, Guangzhou Chest Hospital and First People’s Hospital of Chenzhou. In addition, we used the TCGA-NSCLC dataset to further investigate the mutation characteristics, immune cell infiltration characteristics, immune-related gene expression profiles (GEPs), tumor immunogenicity, and antitumor immunity in patients with MET amplification (Figure S1).
The gene mutation landscape in the ICI-treated cohort (Rizvi et al.) is shown in Figure 1A, along with information for other genes mutated in the MET-amplification group (1.67%, 4/240), such as TP53 (75%, 3/4), KEAP1 (50%, 2/4), SMARCA4 (50%, 2/4), PTPRT (50%, 2/4), EGFR (25%, 1/4), and PTPRD (25%, 1/4). Most were missense mutations (6/11, 54.5%) and frameshift mutations (3/11, 27.3%). The gene mutation landscape in the non-ICI-treated cohort (TCGA-NSCLC) is shown in Figure 1B. In the MET-amplification group (2.58%, 24/929), the TP53 (70.83%, 17/24), TTN (50%, 12/24), CSMD3 (45.83%, 11/24), LRP1B (41.67%, 10/24), ZFHX4 (41.67%, 10/24), XIRP2 (37.5%, 9/24), COL11A1 (33.33%, 8/24), MUC16 (33.33%, 8/24), PCDH15 (33.33%, 8/24), NAV3 (29.17%, 7/24), and RYR2 (29.17%, 7/24) genes had the highest mutation frequency.
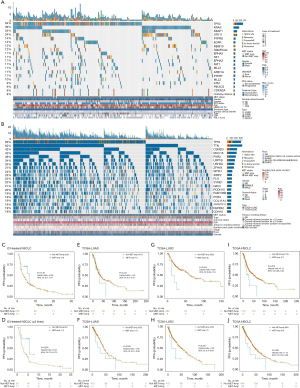
Survival analyses based on the MET status
To further explore the correlation between MET amplification and PFS on immunotherapy in patients with NSCLC, we collected and pooled two ICI-treated cohorts—the Rizvi et al. cohort (n=240) and our local cohort (n=10). In this integrated ICI-treated cohort, the MET-amplification group had a trend toward a longer PFS than the non-MET-amplification group (Figure 1C; P=0.149, HR =0.56, 95% CI: 0.30–1.04). As the local cohort consisted of patients with MET amplification who were treated with ≥3 treatment lines, we adjusted for the treatment lines and further performed the comparison in the integrated ICI-treated cohort restricted to ≥3 treatment lines (n1=62, n2=10). This showed MET amplification was associated with prolonged PFS after immunotherapy (Figure 1D; P=0.039, HR =0.37, 95% CI: 0.18–0.73).
To confirm the efficacy of MET amplification for predicting the PFS and OS of non-ICI-treated patients with NSCLC, we further evaluated the survival difference between patients with MET amplification and those without MET amplification in the TCGA-NSCLC LUAD/LUSC cohort and found no differences in OS or PFS between patients with MET amplification and those without MET amplification (TCGA-NSCLC LUAD/LUSC) (Figure 1E-1J).
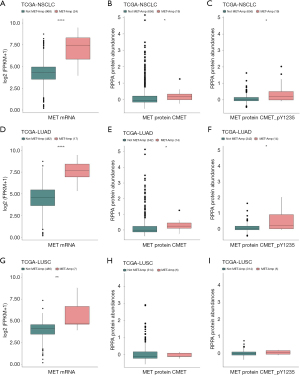
Association of MET gene amplification with upregulated MET mRNA and protein expression
In NSCLC, c-MET gene amplification further leads to abnormal activation of the c-MET pathway via overexpression of the c-MET protein. Abnormal activation of the c-MET pathway ultimately promotes tumor cell proliferation and metastasis (31). We found higher MET mRNA and protein expression in the MET-amplification group than in the non-MET-amplification group in the TCGA-NSCLC LUAD/LUSC cohort (Figure 2A-2I). However, the regulatory effect of MET amplification on the expression of MET mRNA and protein during the evolution of NSCLC is unclear. Our analysis based on the TCGA-NSCLC LUAD/LUSC cohort showed that the MET-amplification NSCLC group (2.41%, 24/992) had higher expression levels of MET mRNA and MET proteins CMET and CMET_PY1235 than the non-MET-amplification group (Figure 2A-2C; P<0.0001, P<0.05, and P<0.05, respectively). Similarly, the MET-amplification LUAD group had higher expression levels of MET mRNA and MET proteins CMET and CMET_PY1235 than the non-MET-amplification LUAD group (Figure 2D-2F; P<0.0001, P<0.05, and P<0.05, respectively). In addition, the MET-amplification LUSC group had higher expression levels of MET mRNA (Figure 2G, P<0.01) and a trend toward higher expression levels of the MET proteins CMET and CMET_PY1235 (Figure 2H,2I) than the non-MET-amplification LUAD group.
Most current treatment strategies for abnormal activation of the c-MET pathway use multikinase MET inhibitors such as crizotinib and cabozantinib. The GDSC database contains data for approximately 1,000 human cancer cell lines. Regarding the susceptibility to different drugs, we further investigated the correlation between MET amplification and the sensitivity to common drugs in 69 NSCLC cell lines from the GDSC database (Figure S2). There was no significant difference in common chemotherapeutic drugs (cisplatin, paclitaxel, docetaxel, vinorelbine, and gemcitabine) and c-MET receptor kinase inhibitors (cabozantinib; crizotinib; foretinib; and PHA-665752), between the MET-amplification and non-MET-amplification NSCLC groups. In addition, NSCLC cell lines with MET amplification were resistant to common targeted drugs, such as c-MET receptor kinase inhibitors (e.g., crizotinib). These drug sensitivity data showed that MET-amplified NSCLC may be less responsive to chemotherapy or targeted therapy.
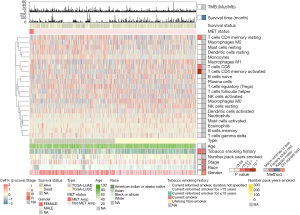
Landscape of immune cells and transcriptome traits based on MET status
To further investigate differences in the immune cell infiltration status and potential biological mechanisms between NSCLC with and without MET amplification, we used the CIBERSORT algorithm to estimate the immune cell infiltration status in 992 samples from the TCGA-NSCLC cohort (Figure 3). The heatmap indicated that the immune cells and some clinical features were significantly different between the MET-amplification and non-MET-amplification groups. For example, CD8+ T cells (P<0.05), M1 macrophages (P<0.05), gamma delta T cells (P<0.05), and activated CD4+ memory T cells (P<0.0001) were more abundant in the MET-amplification group. This pattern indicated that immune-activated cells were significantly enriched in the MET-amplification group. In addition, patients in the MET-amplification group had a longer duration of smoking (median: MET amplification vs. non-MET amplification: 45 vs. 30 years, P<0.05) and were more likely to have a history of smoking (P<0.01).
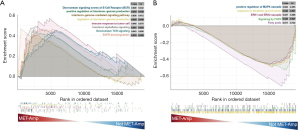
As MET amplification plays a key role in NSCLC progression and prognosis, we analyzed the underlying biological mechanisms of MET amplification in the TCGA-NSCLC dataset. The TCGA-NSCLC dataset was divided into two groups according to the MET amplification status, and GSEA was performed using the gene set obtained from the MSigDB. When the MET-amplification group was compared with the non-MET-amplification group, immune response-related pathways, such as downstream signaling events of B cell receptors (BCRs), positive regulation of interferon-gamma (IFN-γ) production, immune response to tumor cells, and downstream TCR signaling, were significantly upregulated in the MET-amplification group (Figure 4A). Additionally, oncogenic signaling and metabolic-related pathways, such as response to fibroblast growth factor, ERK1 and ERK2 cascade, and fatty acids, were significantly downregulated in the MET-amplification group (Figure 4B).
Association of MET gene amplification with enhanced immunogenicity and activated antitumor immunity
To better identify the immune profile, we analyzed differences in immune-related gene expression patterns between the MET-amplification and non-MET-amplification groups in the TCGA-NSCLC database (Figure 5A-5D). Consistent with the elevated immune cell infiltration, the expression of many immunostimulation-related genes, such as chemokine genes (CXCL9, CXCL10, and CCL5) and cytolysis-related genes (PRF1 and GZMA), was significantly upregulated in the MET-amplification group (Figure 5A,5B). The MET-amplification NSCLC group exhibited significant upregulation of immune checkpoint-related genes (such as LAG3 and PDCD1LG2) compared to the non-MET-amplification NSCLC group. In addition, the MET-amplification NSCLC group exhibited significantly upregulated expression of antigen presentation-related genes, such as HLA-A, HLA-B, HLA-C, and MICB (Figure 5A). Further subgroup analysis showed that the MET-amplification TCGA-LUAD group also had upregulated immune-related gene expression patterns such as chemokine-related, cytotoxicity-related and immune checkpoint-related genes (Figure 5A,5B). These results suggest that MET amplification is closely associated with enhanced tumor immunogenicity and an activated immune microenvironment, providing a theoretical basis for the benefit of immunotherapy in patients with MET-amplified NSCLC.
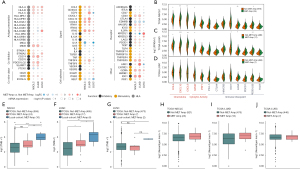
We further compared tumor immunogenicity and antitumor immunity between patients with and without MET amplification in the TCGA-NSCLC cohort. Figure 5E shows that 10 patients with MET amplification (local cohort, ICI-treated) had a higher TMB than those without (TCGA-NSCLC, non-ICI-treated; P<0.0001), whereas all patients with MET amplification had a trend toward a higher TMB than those without (TCGA-NSCLC, non-ICI-treated; P<0.01). Subgroup analysis (Figure 5F) showed that the MET-amplification LUAD group (local cohort) had a higher TMB than the non-MET-amplification (P<0.001) and MET-amplification (P<0.05) TCGA-LUAD groups, whereas the MET-amplification TCGA-LUAD group also had a higher TMB than the non-MET-amplification TCGA-LUAD group (P<0.05). Similarly, the MET-amplification LUSC group (local cohort) had a higher TMB than the non-MET-amplification TCGA-LUSC group (P<0.05, Figure 5G). In addition, the MET-amplification TCGA-NSCLC group had a trend toward a higher NAL than the non-MET-amplification TCGA-NSCLC group (Figure 5H). Further subgroup analysis showed that the levels of NAL between the MET-amplification and non-MET-amplification in the TCGA-LUAD group (Figure 5I) and TCGA-LUSC group (Figure 5J).
Increasingly, studies have shown that alterations in DNA damage response- or DNA damage repair-related pathways are associated with immunotherapeutic efficacy (32-34). For example, the number of DDR pathway gene mutations is positively correlated with markers of tumor immunogenicity, such as TMB and NAL. In addition, DDR may be a biomarker for predicting the efficacy of immunotherapy (32). Therefore, we used the DDR gene set from the MSigDB (https://cdn.amegroups.cn/static/public/atm-21-4543-2.xlsx) to compare differences in the number of mutations in the DDR-related pathway between the MET-amplification and non-MET-amplification groups in the TCGA-NSCLC/LUAD/LUSC cohort and in GDSC-NSCLC cell lines (Figure S3). The number of mutations in multiple DDR pathways was significantly higher in the MET-amplification TCGA-NSCLC LUAD group than in the non-MET-amplification TCGA-NSCLC LUAD group.
Discussion
MET amplification plays critical roles in the NSCLC mutagenic process, contributing to subclonal diversification, intratumor heterogeneity. Here, our study focused on the association between MET amplification and the efficacy of ICIs for NSCLC treatment, and the results showed it was associated with superior PFS times in the ICI-treated cohort (n=72; P=0.039, HR =0.37, 95% CI: 0.18–0.73) but not in the non-ICI-treated cohort (e.g., the TCGA-NSCLC LUAD/LUSC cohort). Thus, MET amplification can distinctly function in NSCLC to predict the prognosis of immunotherapy. Through analyses of immune cells, immune-related gene expression, immunogenicity, and antitumor immunity, we identified significant correlations between MET amplification and immune-related gene expression, increased immune cell infiltration, enhanced immunogenicity, activated antitumor immunity, and decreased DDR efficacy. In addition, in the GDSC database, the MET-amplification group exhibited a trend toward resistance to many common chemotherapeutic drugs, such as c-MET receptor kinases (e.g., crizotinib). Studies have shown that patients with MET overexpression may be more responsive to ICI immunotherapy than to targeted therapy (15), and MET amplification, mutation, or overexpression may be manifested by upregulation of PD-L1 and other immunosuppressive molecules at the mRNA and protein levels (17). Furthermore, we summarized the possible mechanisms underlying the improved efficacy and prognosis in MET-amplified NSCLC patients receiving ICIs (Figure 6).
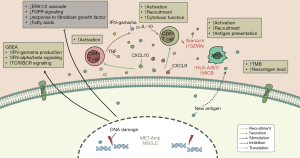
Tumor immunogenicity is the developmental basis of tumor immunity, and many somatic mutations can produce antigens to activate CD8+ cytotoxic T cells, thereby exerting a T cell-mediated antitumor effect. In addition, IFN-γ is generally produced by effector T cells or antigen-presenting cells (APC) to support antigen presentation and the recruitment of other immune cells, thereby initiating antiproliferative and apoptotic effects on the tumor. To date, T cell-inflamed GEPs, specific gene mutations, high MSI (MSI-H), TMB, NAL, and the TME (such as the CD8+ T cell abundance) have gradually become potential markers for the immunotherapy response (3,10,35). When TMB increases, it promotes the production of new antigens in tumors which are presented to APC, which can lead to the transformation of T cells into mature and activated T cells and increase the sensitivity of patients to treatment with PD-1 and CTLA-4 inhibitors (36). Recently, mutations in DDR pathway-related genes have attracted attention in immunotherapy (37). For example, functional mutations in the DDR pathway reduced genome stability and increased tumor immunogenicity via the accumulation of DNA damage to increase the efficacy of immunotherapy (37,38). This effect suggests ICI therapy may be a potential strategy for patients with MET-amplified NSCLC. However, no clinical correlation has been identified for MET immunotherapy in NSCLC.
As is known to all, MET amplification is a de novo driver gene in NSCLC about 5%. So far, there are several c-MET inhibitors approved by FDA/CFDA for MET exon 14 skipping in advanced NSCLC. However, for de novo MET amplification, MET-TKI monotherapy demonstrated a short PFS of 4 months in NSCLC clinical study. In a subgroup analysis of PROFILE 1001, patients with high level MET amplification demonstrated an objective response rate (ORR) of 38.1% and a long PFS of 6.7 months. These result suggested MET inhibitors maybe only efficacy to patients with high level MET amplification in NSCLC. And our study demonstrated immunotherapy maybe another selective strategy for patients with low level MET amplification. On the other side, MET amplification is an acquired resistance gene closed to 20% after using EGFR-TKI. In a phase I/II clinical study “INSIGHT”, tepotinib combined with gefitinib demonstrated a dramatic PFS of 16.6 months which provided a new therapy for patients with resistance from EGFR-TKI, especially from third EGFR-TKI in future. In this study we found MET gene amplification was associated with inflammatory tumor microenvironment, but whether it can remodel the immune desert type of tumor microenvironment in EGFR-mutant NSCLC has not been known. Whether these patients can be benefit from immunotherapy still need further research.
Different from target therapy and chemotherapy, immunotherapy in advanced NSCLC is focus on the long-term survival. Existing studies suggest that RECIST 1.1 evaluation underestimated the benefit of immunotherapy, further research is required to optimize iRECIST and establish some criteria for selecting patients who will benefit from continued immunotherapy beyond PD per RECIST 1.1. Tumor micrometastasis maybe one of key point about recurrence after lung cancer surgery. It was reported that tumor immune microenvironment is predictive of prognosis after surgery in NSCLC. This suggested immunotherapy may impact on tumor micrometastasis and improve the prognosis of early lung cancer. Lambrechts et al. (39) reported that the expression profiles of tumor stromal cell marker genes and tumor stromal cell subsets differed between LUAD and LUSC, and that low expression of CD8+ T cell cluster marker genes in LUAD was associated with an improved survival prognosis. In contrast, CD8+ T cell cluster marker genes expressed in LUSC were associated with a worse survival prognosis in LUSC. All these studies suggested some special patients maybe benefit from immunotherapy after surgery. To date, clinical research about evaluation of immunotherapy after lung cancer surgery are going on. In this study, we found MET amplification is closely associated with enhanced tumor immunogenicity and an activated immune microenvironment. Therefore, it worth waiting for whether patients with MET amplification after surgery can benefit from checkpoint inhibitors.
This study has some limitations. First, our analysis did not compare MET amplification with non-MET amplification in patients with NSCLC on first-line immunotherapy. Second, unlike the NSCLC ICI-treated cohort from Rizvi et al., our local datasets represent only the Chinese population, and differences in genetic backgrounds between ethnicities may affect the results of the analysis. Third, we found patients in the MET-amplification NSCLC group (local cohort) had a higher TMB than those in the MET-amplification TCGA-NSCLC group, which showed, TMB may be a predictive marker of the clinical immunotherapy response in the MET-amplification NSCLC group (local cohort) to select patients for immunotherapy. Fourth, MET amplification has been reported to show intratumoral heterogeneity in a number of cases (40), and intratumoral heterogeneity should be considered when interpreting our results on MET and phospho-MET protein expression. Fifth, the threshold used to define MET amplification varies among studies. Sixth, because of the difference in structure, there still a certain difference in the efficacy of different ICIs. But in this study, the number of patients treated with ICIs was unfortunately very small. So we cannot to explore it further. Finally, our analysis considered only the two most important subtypes of NSCLC, and the remaining subtypes were not considered. Therefore, more research involving a large number of samples and diverse ethnic groups is needed for analysis and validation.
Conclusions
Our study provides evidence that MET amplification is associated with long PFS times and with known immunotherapy response markers, including TMB, NAL, immune-related genes, and the high infiltration of specific immune cells. Therefore, MET amplification could be a predictive biomarker for ICIs. A series of prospective clinical studies and molecular mechanistic explorations is required.
Acknowledgments
We thank the biotrainee, Dr. Jianming Zeng (University of Macau); XiaoYa HuaTu, Lianchuan Biotechnology Limited (subsidiary of LC Sciences, Hangzhou, China). A part of data had previously accepted as an abstract in the “20th IASLC Word Conference on Lung Cancer”.
Funding: This work was supported by the National Natural Science Foundation of China (82003304) and Guangzhou Science and Technology Program, China (202102080013).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-4543
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-4543
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-4543). Dr. JZ reports support from Haplox Biotechnology Co., Ltd. The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This research was performed in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the ethics committee of Zhujiang Hospital of Southern Medical University, Guangzhou Chest Hospital and First People’s Hospital of Chenzhou. Written informed consent was obtained from the individuals for the publication of any potentially identifiable images or data included in this article.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019;19:133-50. [Crossref] [PubMed]
- Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018-28. [Crossref] [PubMed]
- Lin A, Wei T, Meng H, et al. Role of the dynamic tumor microenvironment in controversies regarding immune checkpoint inhibitors for the treatment of non-small cell lung cancer (NSCLC) with EGFR mutations. Mol Cancer 2019;18:139. [Crossref] [PubMed]
- Mehnert JM, Panda A, Zhong H, et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Invest 2016;126:2334-40. [Crossref] [PubMed]
- Riaz N, Havel JJ, Kendall SM, et al. Recurrent SERPINB3 and SERPINB4 mutations in patients who respond to anti-CTLA4 immunotherapy. Nat Genet 2016;48:1327-9. [Crossref] [PubMed]
- Shin DS, Zaretsky JM, Escuin-Ordinas H, et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov 2017;7:188-201. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Boeri M, Milione M, Proto C, et al. Circulating miRNAs and PD-L1 tumor expression are associated with survival in advanced NSCLC patients treated with immunotherapy: a prospective study. Clin Cancer Res 2019;25:2166-73. [Crossref] [PubMed]
- Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593 [Crossref] [PubMed]
- Marcus L, Lemery SJ, Keegan P, et al. FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin Cancer Res 2019;25:3753-8. [Crossref] [PubMed]
- Schoenfeld AJ, Rizvi H, Bandlamudi C, et al. Clinical and molecular correlates of PD-L1 expression in patients with lung adenocarcinomas. Ann Oncol 2020;31:599-608. [Crossref] [PubMed]
- Wu HX, Chen YX, Wang ZX, et al. Alteration in TET1 as potential biomarker for immune checkpoint blockade in multiple cancers. J Immunother Cancer 2019;7:264. [Crossref] [PubMed]
- Guisier F, Dubos-Arvis C, Viñas F, et al. Efficacy and safety of anti-PD-1 immunotherapy in patients with advanced NSCLC with BRAF, HER2, or MET mutations or RET translocation: GFPC 01-2018. J Thorac Oncol 2020;15:628-36. [Crossref] [PubMed]
- Mazieres J, Cropet C, Montané L, et al. Vemurafenib in non-small-cell lung cancer patients with BRAFV600 and BRAFnonV600 mutations. Ann Oncol 2020;31:289-94. [Crossref] [PubMed]
- Reis H, Metzenmacher M, Goetz M, et al. MET expression in advanced non-small-cell lung cancer: effect on clinical outcomes of chemotherapy, targeted therapy, and immunotherapy. Clin Lung Cancer 2018;19:e441-63. [Crossref] [PubMed]
- Westover D, Zugazagoitia J, Cho BC, et al. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann Oncol 2018;29:i10-9. [Crossref] [PubMed]
- Ahn HK, Kim S, Kwon D, et al. MET receptor tyrosine kinase regulates the expression of co-stimulatory and co-inhibitory molecules in tumor cells and contributes to PD-L1-mediated suppression of immune cell function. Int J Mol Sci 2019;20:4287. [Crossref] [PubMed]
- Sabari JK, Leonardi GC, Shu CA, et al. PD-L1 expression, tumor mutational burden, and response to immunotherapy in patients with MET exon 14 altered lung cancers. Ann Oncol 2018;29:2085-91. [Crossref] [PubMed]
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol 2018;36:633-41. [Crossref] [PubMed]
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. [Crossref] [PubMed]
- Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016;44:e71 [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2013;41:D955-61. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity 2018;48:812-30.e14. [Crossref] [PubMed]
- Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 2017;377:2500-1. [Crossref] [PubMed]
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847-9. [Crossref] [PubMed]
- Organ SL, Tsao MS. An overview of the c-MET signaling pathway. Ther Adv Med Oncol 2011;3:S7-S19. [Crossref] [PubMed]
- Wang Z, Zhao J, Wang G, et al. Comutations in DNA damage response pathways serve as potential biomarkers for immune checkpoint blockade. Cancer Res 2018;78:6486-96. [Crossref] [PubMed]
- Teo MY, Seier K, Ostrovnaya I, et al. Alterations in DNA damage response and repair genes as potential marker of clinical benefit from PD-1/PD-L1 blockade in advanced urothelial cancers. J Clin Oncol 2018;36:1685-94. [Crossref] [PubMed]
- Luo P, Lin A, Li K, et al. DDR pathway alteration, tumor mutation burden, and cisplatin sensitivity in small cell lung cancer: difference detected by whole exome and targeted gene sequencing. J Thorac Oncol 2019;14:e276-9. [Crossref] [PubMed]
- Ott PA, Bang YJ, Piha-Paul SA, et al. T-cell-inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 cancers: KEYNOTE-028. J Clin Oncol 2019;37:318-27. [Crossref] [PubMed]
- McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463-9. [Crossref] [PubMed]
- Ricciuti B, Recondo G, Spurr LF, et al. Impact of DNA damage response and repair (DDR) gene mutations on efficacy of PD-(L)1 immune checkpoint inhibition in non-small cell lung cancer. Clin Cancer Res 2020;26:4135-42. [Crossref] [PubMed]
- Park S, Lee H, Lee B, et al. DNA damage response and repair pathway alteration and its association with tumor mutation burden and platinum-based chemotherapy in SCLC. J Thorac Oncol 2019;14:1640-50. [Crossref] [PubMed]
- Lambrechts D, Wauters E, Boeckx B, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med 2018;24:1277-89. [Crossref] [PubMed]
- Yoshimura K, Karayama M, Inoue Y, et al. Heterogeneous MET gene copy number and EGFR mutation elicit discordant responses to crizotinib between primary and metastatic lesions in erlotinib-resistant lung adenocarcinoma. Lung Cancer 2018;124:317-9. [Crossref] [PubMed]
(English Language Editor: B. Draper)


