
Successful targeting of the NRG1 fusion reveals durable response to afatinib in lung adenocarcinoma: a case report
Introduction
Oncogenic gene fusions act as actionable drivers in the initiation and development of solid cancers (1). One of the actionable gene fusions involves the neuregulin 1 gene (NRG1), which was recently identified as recurrent aberrations in patients with non-small cell lung cancer (NSCLC) (2,3). The prevalence of NRG1 fusion in lung cancer was relatively low compared with other common driver mutations. In a study, NRG1 fusions were identified in 25 of 9,592 NSCLC cases (0.26%) (4). Previous studies have suggested that fusions involving NRG1 could lead to the activation of the erythroblastic leukemia viral oncogene homolog (ErbB)-mediated pathway and could therefore serve as a therapeutic candidate in addition to currently available ErbB-targeted treatment (5,6). The irreversible pan-ErbB inhibitor, afatinib, has been approved for NSCLC patients with EGFR sensitizing mutations including L858R and exon 19 deletions (7). However, clinical evidence on treatment effects of EGFR targeted therapies such as afatinib in NRG1 fusion-positive lung cancer is limited due to relatively low frequency of NRG1 fusions. Previous studies reported the use of afatinib as front-line or later-line treatments in NSCLC patients with NRG1 fusions (8,9).
In the present study, we reported an elder Chinese patient with lung adenocarcinoma harboring the NRG1 fusion. The patient received afatinib as first-line treatment. After afatinib resistance, our study performed additional next-generation sequencing (NGS) test on patient sample and tried to identify underlying drug resistant mechanisms. We present the following case in accordance with the CARE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3923).
Case presentation
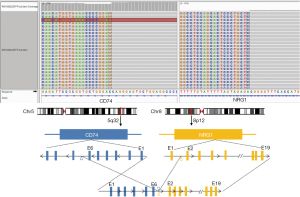
An 80-year-old female, Chinese, non-smoker was admitted to the First Affiliated Hospital of Wenzhou Medical University due to sharp pain in her left chest in October 2018. Computed tomography (CT) scan (Figure 1) of the chest displayed a 44 mm × 41 mm nodular lesion in the left upper lung (Figure 1A). The patient was diagnosed with invasive adenocarcinoma with bone metastasis. Both lung biopsy and plasma samples were subjected to the next-generation sequencing (NGS) platform with a panel of 168 solid cancer-related genes (Burning Rock Biotech, Guangzhou, China). CD molecule 74 (CD74)-NRG1 fusion was identified in both the biopsy and plasma samples (Figure 2), and the co-mutation of tumor protein p53 (TP53) was also indicated in the biopsy sample result.
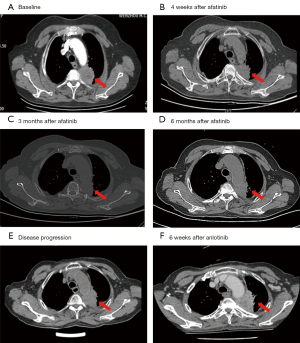
The patient was started on 40 mg afatinib orally daily from November 2018. After 4 weeks of afatinib treatment, the chest CT scan displayed tumor diminution, and the target lesion size significantly decreased to 28 mm × 24 mm (Figure 1B). The patient achieved partial response (PR). Side effects including weight loss, acute cough, mild rash, and diarrhea were observed and controlled during the treatment. The tumor decreased to 27 mm × 23 mm 3 months after treatment (Figure 1C). Treatment-response assessment indicated the patient remained in PR, with a slight growth to 29 mm × 27 mm in May 2019 (Figure 1D). In July 2019, a chest CT scan showed significant growth of the left upper lobe (37 mm × 34 mm), and the patient was diagnosed with progressive disease (PD) (Figure 1E). Progression-free survival (PFS) was 8 months. The plasma sample obtained after PD was sent for NGS in August 2019, but no new mutations were detected compared with the previous test.
The second-line treatment was switched to anlotinib in September 2019. The patient reported gastrointestinal side-effects and remained in stable disease (SD) for 6 weeks. The patient had PD in November 2019 (Figure 1F), with 2 months’ PFS.
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Discussion
Identifying gene rearrangements as novel oncogenic drivers can improve the standard of care for advanced NSCLC treatment. There have been increasing investigations on oncogenic gene fusions, while some rare gene fusions were proposed as therapeutic targets of tyrosine kinase inhibitors (1).
Recent studies reported the presence of NRG1 rearrangements in different cancer types (3). The most frequently reported NRG1 fusion is CD74-NRG1, while the expression of this fusion protein-induced signaling through ErbB2–ErbB3 heterodimers (10). Therefore, patients harboring the CD74-NRG1 fusion may respond to pan-ErbB inhibitors, such as afatinib. Our patient harboring the NRG1 fusion showed a durable and significant response to afatinib, with a PFS of 8 months. Our case further supported the use of ErbB-targeted therapy in the patient positive for NRG1 fusion.
Fusions involving NRG1 appear to have a low incidence in solid tumors (10). The oncogenic mechanism underlying NRG1 fusions and encouraging results from previous clinical studies indicated the use of TKI as a potential treatment option. The NRG1 gene contains large intronic regions, which have been recognized as a challenge in detecting complex and divers NRG1 fusions. In our case, NGS showed the clinical adaption for the detection of low-incidence mutations, such as NRG1 gene rearrangement. Therefore, our case supported the use of NGS in detecting NRG1 fusions.
Although the patient showed a significant therapeutic response to afatinib, acquired drug resistance inevitably occurred. The second NGS test did not identify novel mutations that emerged after afatinib resistance. More comprehensive studies at proteome and metabolome levels may improve our understanding of the mechanism for acquired afatinib resistance.
Acknowledgments
We want to thank the patient and her family for their contributions to the study.
Funding: This work was funded by the Zhejiang Province Public Welfare Technology Application Research Project (No. 2017C33022) and the Zhejiang Natural Science Foundation (No. LY19H160023).
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3923
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3923). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Schram AM, Chang MT, Jonsson P, et al. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol 2017;14:735-48. [Crossref] [PubMed]
- Latysheva NS, Babu MM. Discovering and understanding oncogenic gene fusions through data intensive computational approaches. Nucleic Acids Res 2016;44:4487-503. [Crossref] [PubMed]
- Fernandez-Cuesta L, Thomas RK. Molecular Pathways: Targeting NRG1 Fusions in Lung Cancer. Clin Cancer Res 2015;21:1989-94. [Crossref] [PubMed]
- Jonna S, Feldman RA, Swensen J, et al. Detection of NRG1 Gene Fusions in Solid Tumors. Clin Cancer Res 2019;25:4966-72. [Crossref] [PubMed]
- Riely GJ, Yu HA. EGFR: The Paradigm of an Oncogene-Driven Lung Cancer. Clin Cancer Res 2015;21:2221-6. [Crossref] [PubMed]
- Drilon A, Somwar R, Mangatt BP, et al. Response to ERBB3-Directed Targeted Therapy in NRG1-Rearranged Cancers. Cancer Discov 2018;8:686-95. [Crossref] [PubMed]
- Liao BC, Lin CC, Yang JC. Novel EGFR Inhibitors in Non-small Cell Lung Cancer: Current Status of Afatinib. Curr Oncol Rep 2017;19:4. [Crossref] [PubMed]
- Gay ND, Wang Y, Beadling C, et al. Durable Response to Afatinib in Lung Adenocarcinoma Harboring NRG1 Gene Fusions. J Thorac Oncol 2017;12:e107-e110. [Crossref] [PubMed]
- Cadranel J, Liu SV, Duruisseaux M, et al. Therapeutic Potential of Afatinib in NRG1 Fusion-Driven Solid Tumors: A Case Series. Oncologist 2021;26:7-16. [Crossref] [PubMed]
- Fernandez-Cuesta L, Plenker D, Osada H, et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discov 2014;4:415-22. [Crossref] [PubMed]
(English Language Editors: R. Scott and J. Chapnick)


