
Soluble LRIG2 is a potential biomarker for type 2 diabetes mellitus
Introduction
Type 2 diabetes mellitus is a major public health problem worldwide. According to the International Diabetes Federation (IDF), the global prevalence of diabetes in 2019 was 9.3% (approximately 463 million adults), and is projected to increase to 10.9% (700 million adults) by 2045. However, around half (50.1%) of all individuals with diabetes are undiagnosed (1). Type 2 diabetes undiagnosed for many years is associated with an increased risk of microvascular and macrovascular complications (2,3). Early diagnosis and effective treatment can delay the onset of diabetic complications. The current gold standard for diagnosing diabetes is the oral glucose tolerance test (OGTT); however, due to its complexity, fasting plasma glucose is mainly used for screening in health checkups, often leading to misdiagnosis. Therefore, a novel biomarker for diabetes is needed.
The leucine-rich repeats and immunoglobulin-like domains (LRIG) gene family, consisting of LRIG1–3, encodes a transmembrane protein and regulates growth factor receptor signaling (4). LRIG1 is a negative regulator of several receptor tyrosine kinases, including the erythroblastic leukemia viral oncogene homolog (ErbB) family, and has been identified as a tumor suppressor in various cancers (5,6). Conversely, LRIG 2 positively regulates the epidermal growth factor receptor (EGFR) signaling pathway, and its expression is associated with poor survival in several cancers (7,8). LRIG3 is less studied than LRIG1 and LRIG2, and its function remains unclear. Recently, LRIG3 was proposed to oppose LRIG1 (9); however, other studies indicated that LRIG3 negatively regulates EGFR signaling (10,11). The EGFR pathway plays a key role in tumorigenesis, and dysregulation of the pathway is common in various human cancers (12,13).
In several studies, alteration of the EGFR pathway was shown to affect glucose metabolism and inhibition of EGFR improved insulin resistance in type 2 diabetes (14,15). Miettinen et al. reported that EGFR plays an important role in pancreatic beta cell mass regulation; downregulation of EGFR signaling pancreatic islets caused diabetes in a mouse model (16,17). We previously showed that serum soluble EGFR (sEGFR) levels are higher in newly diagnosed type 2 diabetic patients than in controls (18). There has been very little research that explores the role of LRIG in metabolism. However, a recent study indicated the association between LRIG1 variant and risk of type 2 diabetes (19). In the present study, the usefulness of serum soluble LRIG2 (sLRIG2) as an EGFR-related diagnostic biomarker of type 2 diabetes was evaluated. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/atm-21-3272).
Methods
Study design
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the Institutional Review Board of Chungnam National University Hospital (CNUH 2014-12-013) and written informed consent was obtained from each participant before enrollment. A total of 240 subjects with newly diagnosed type 2 diabetes (n=80), prediabetes (n=80), or normal glucose tolerance (NGT; n=80) were recruited from the Department of Endocrinology of Chungnam National University Hospital. Subjects who used oral hypoglycemic agents or tyrosine kinase inhibitors (TKIs) were excluded. Subjects with acute infectious disease or a history of malignancy were also excluded. Medical history (age, sex, drug use, smoking, underlying disease) and anthropometric measurements (height and body weight) were taken. Plasma samples were obtained from all subjects for the measurement of biochemical markers at baseline and 2 hours after ingestion of a 75-g oral glucose load. Type 2 diabetes was diagnosed according to the criteria of the American Diabetes Association (ADA, Diagnosis and classification of diabetes mellitus, 2019). Prediabetes was defined as fasting plasma glucose 100 to 125 mg/dL or 2-hour postprandial glucose 140 to 199 mg/dL or hemoglobin A1C (HbA1c) 5.7% to 6.4% according to the ADA criteria.
Biochemical data
Blood samples were collected using ethylenediaminetetraacetic acid tubes in the morning after an overnight fast of more than 8 hours, and 2 hours after ingestion of a 75-g oral glucose load. The serum samples were obtained by centrifuging the blood. Lipids, including high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), total cholesterol (TC), and triglycerides (TGs), were measured using a blood chemistry analyzer (Hitachi 747; Hitachi, Tokyo, Japan). Insulin was quantified using an immunoradiometric assay kit (INS-IRMA Kit; DIAsource, Louvain-la-Neuve, Belgium). HbA1c was measured using high-performance liquid chromatography (Bio-Rad, Hercules, CA, USA). The homeostasis model assessment-estimated insulin resistance (HOMA-IR) was calculated as the fasting insulin (µU/mL) × fasting glucose (mmol/L)/22.5. The homeostasis model assessment of β-cell function (HOMA-β) was calculated using the following formula: 20 × fasting insulin (µU/mL)/fasting glucose (mmol/L) − 3.5 (20).
The fasting serum sLRIG2 level was measured using a quantitative sandwich enzyme immunoassay technique with an enzyme-linked immunosorbent assay (ELISA) kit (MyBioSource, San Diego, CA, USA; catalog number MBS9337066, intra-assay precision <15%, inter-assay precision <15%).
Statistical analyses
All statistical analyses were performed using SPSS statistical software for Windows (version 21.0; SPSS Inc., Chicago, IL, USA) and GraphPad Prism software (version 7.0; GraphPad Software Inc., La Jolla, CA, USA). Continuous variables are presented as the mean ± standard deviation. Differences were considered statistically significant at P<0.05. Baseline demographics including biochemical data were compared using one-way analysis of variance (ANOVA) with Bonferroni correction and the chi-square test. Analyses of covariance (ANCOVA) were also performed to control for the effects of covariates. Pearson’s correlation and linear regression analyses were used to investigate the correlations between sLRIG2 and various clinical factors. To evaluate the sensitivity and specificity of serum sLRIG2 in the diagnosis of type 2 diabetes, the area under the receiver operating characteristic (ROC) curve analysis was performed.
Results
Demographics
The clinical characteristics of the study participants are presented in Table 1. Significant differences were observed in body mass index, body weight, glucose (fasting, 2-hour postprandial), C-peptide (fasting), insulin (fasting, 2-hour postprandial), HbA1c, HOMA-IR, TG, HDL-C, aspartate aminotransferase (AST), and alanine aminotransferase (ALT) among the three groups. In addition, hypertension and current smoking status were more common in the type 2 diabetes group. Age, sex, height, blood pressure (systolic and diastolic), C-peptide (2-hour postprandial), HOMA-β, TC, LDL-C, blood urea nitrogen, creatinine, and the estimated glomerular filtration rate were not different among the three groups.
Table 1
| Variable | NGT (n=80) | Prediabetes (n=80) | T2DM (n=80) | P value |
|---|---|---|---|---|
| Age, mean ± SD (years) | 50.7±13.7 | 53.5±12.1 | 54±12.3 | 0.210* |
| Sex (male/female) | 25/55 | 38/42 | 35/45 | 0.091† |
| Hypertension, n (%) | 16 (20.0) | 14 (17.5) | 33 (41.3) | 0.001† |
| Smoking, n (%) | 8 (10.0) | 7 (8.8) | 22 (27.5) | 0.001† |
| Height, mean ± SD (cm) | 161.6±8.6 | 163.2±8.4 | 163.3±9.4 | 0.528* |
| Weight, mean ± SD (kg) | 63.9±12.4 | 67.1±11.9 | 70.1±14.7 | 0.039* |
| BMI, mean ± SD (kg/m2) | 24.3±3.3 | 24.8±3.2 | 26.2±4.4 | 0.020* |
| Systolic BP, mean ± SD (mmHg) | 127.8±16.1 | 128.9±14.6 | 134.6±20.2 | 0.065* |
| Diastolic BP, mean ± SD (mmHg) | 79.3±12.6 | 77.4±10.9 | 81.8±11.8 | 0.102* |
| Hemoglobin, mean ± SD (g/dL) | 13.7±1.2 | 14.1±1.7 | 14.5±1.5 | 0.015* |
| HbA1c, mean ± SD (%) | 5.3±0.2 | 5.6±0.3 | 7.4±1.9 | <0.001* |
| Glucose, fasting, mean ± SD (mg/dL) | 91.9±5 | 101.7±11.1 | 154.2±57.3 | <0.001* |
| Glucose, PP2, mean ± SD (mg/dL) | 105.3±19 | 143.3±33.5 | 293±101.2 | <0.001* |
| Insulin, fasting, mean ± SD (mg/dL) | 8.3±4.2 | 10±5.0 | 14.0±8.5 | <0.001* |
| Insulin, PP2, mean ± SD (mg/dL) | 36.6±43.6 | 68.9±68 | 70.7±69.5 | 0.001* |
| C-peptide, fasting, mean ± SD (ng/mL) | 0.73±0.47 | 0.87±0.5 | 1.15±0.71 | <0.001* |
| C-peptide, PP2, mean ± SD (ng/mL) | 3.44±2.23 | 4.09±2.7 | 6.72±24.34 | 0.347* |
| HOMA-IR, mean ± SD | 1.89±0.99 | 2.52±1.89 | 5.27±3.63 | <0.001* |
| HOMA-β, mean ± SD | 26.04±13.05 | 28.78±15.16 | 29.08±18.38 | 0.409* |
| TG, mean ± SD (mg/dL) | 104±63.2 | 145.1±106.7 | 189.6±173 | <0.001* |
| TC, mean ± SD (mg/dL) | 191.4±30.4 | 196.6±39.8 | 192.3±46.9 | 0.682* |
| HDL-C, mean ± SD (mg/dL) | 61.7±13.7 | 53.7±14 | 49±13.4 | <0.001* |
| LDL-C, mean ± SD (mg/dL) | 113.5±29.7 | 121.5±34.2 | 116.6±45.3 | 0.389* |
| BUN, mean ± SD (mg/dL) | 13.7±3.6 | 14±3.7 | 14.8±4.6 | 0.206* |
| Creatinine, mean ± SD (mg/dL) | 0.73±0.15 | 0.75±0.18 | 0.72±0.19 | 0.496* |
| eGFR, mean ± SD (mL/min/1.73 m2) | 103.4±18.1 | 104.2±24.4 | 112±29.4 | 0.061* |
| AST, mean ± SD (U/L) | 21.3±6.6 | 23.2±11.4 | 26.5±14.9 | 0.019* |
| ALT, mean ± SD (U/L) | 20.7±15 | 25±20.8 | 29.1±16.8 | 0.017* |
*, P value was obtained from student
Comparison of serum sLRIG2 level among the three groups
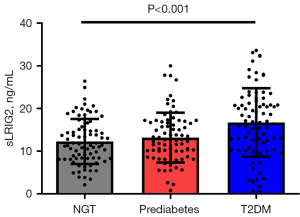
Serum sLRIG2 levels were significantly different among the three groups (P<0.001). In the post-hoc comparisons, serum sLRIG2 levels were significantly higher in subjects with type 2 diabetes (16.7±8.0 ng/mL) than in those without diabetes (NGT group: 12.3±5.3 ng/mL, P<0.001; prediabetes group: 13.2±5.8 ng/mL, P=0.002; Figure 1). Serum sLRIG2 levels were not different between the NGT and prediabetes groups (P=0.999).
Correlation of serum sLRIG2 level with various clinical factors
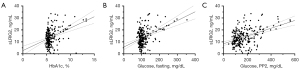
The associations between serum sLRIG2 levels and clinical variables were investigated in all subjects and groups. Body weight, diastolic blood pressure, HbA1c, glucose (fasting, 2-hour postprandial), C-peptide (fasting), and TG significantly correlated with the serum sLRIG2 level in all subjects. Furthermore, height, body weight, HbA1c, and glucose (fasting, 2-hour postprandial) correlated with the serum sLRIG2 level in the type 2 diabetes group (Table 2). HbA1c (r=0.378, P<0.001) and blood glucose (fasting: r=0.421, P<0.001; 2-hour postprandial: r=0.433, P<0.001) correlated more strongly with the sLRIG2 level than other clinical variables (Figure 2).
Table 2
| Variable | Total | NGT | Prediabetes | T2DM | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| r | P | r | P | r | P | r | P | ||||
| Age | −0.065 | 0.317 | 0.122 | 0.280 | −0.160 | 0.157 | −0.211 | 0.060 | |||
| Height | 0.122 | 0.103 | 0.035 | 0.806 | −0.113 | 0.422 | 0.247 | 0.034 | |||
| weight | 0.175 | 0.019 | −0.057 | 0.690 | 0.043 | 0.759 | 0.239 | 0.040 | |||
| BMI | 0.139 | 0.065 | −0.107 | 0.454 | 0.090 | 0.525 | 0.146 | 0.219 | |||
| Systolic BP | 0.179 | 0.016 | 0.270 | 0.055 | −0.088 | 0.528 | 0.185 | 0.114 | |||
| Diastolic BP | 0.216 | 0.004 | 0.236 | 0.096 | 0.153 | 0.269 | 0.197 | 0.092 | |||
| Hemoglobin | 0.109 | 0.128 | −0.006 | 0.962 | 0.006 | 0.966 | 0.110 | 0.396 | |||
| HbA1c | 0.378 | <0.001 | 0.021 | 0.856 | 0.060 | 0.603 | 0.375 | 0.001 | |||
| Glucose, fasting | 0.421 | <0.001 | −0.068 | 0.547 | −0.181 | 0.107 | 0.499 | <0.001 | |||
| Glucose, PP2 | 0.433 | <0.001 | 0.157 | 0.165 | 0.167 | 0.138 | 0.463 | <0.001 | |||
| Insulin, fasting | 0.073 | 0.260 | −0.224 | 0.047 | −0.062 | 0.590 | 0.051 | 0.656 | |||
| Insulin, PP2 | −0.004 | 0.955 | −0.065 | 0.589 | 0.078 | 0.519 | −0.146 | 0.198 | |||
| C-peptide, fasting | 0.144 | 0.034 | 0.014 | 0.909 | 0.090 | 0.453 | 0.061 | 0.603 | |||
| C-peptide, PP2 | −0.001 | 0.988 | 0.107 | 0.376 | 0.136 | 0.259 | −0.062 | 0.606 | |||
| HOMA-IR | 0.188 | 0.004 | −0.219 | 0.052 | −0.109 | 0.343 | 0.119 | 0.294 | |||
| HOMA-β | −0.097 | 0.136 | −0.225 | 0.043 | 0.001 | 0.991 | −0.139 | 0.218 | |||
| TG | 0.176 | 0.007 | 0.070 | 0.540 | −0.001 | 0.995 | 0.168 | 0.153 | |||
| TC | −0.024 | 0.715 | −0.086 | 0.452 | −0.132 | 0.253 | 0.075 | 0.526 | |||
| HDL-C | −0.125 | 0.059 | −0.253 | 0.025 | 0.027 | 0.814 | 0.067 | 0.574 | |||
| LDL-C | −0.074 | 0.264 | 0.003 | 0.978 | −0.164 | 0.154 | −0.071 | 0.555 | |||
| BUN | 0.101 | 0.131 | 0.005 | 0.963 | 0.088 | 0.449 | 0.093 | 0.441 | |||
| Creatinine | 0.096 | 0.152 | 0.096 | 0.401 | 0.087 | 0.453 | 0.145 | 0.228 | |||
| eGFR | 0.033 | 0.623 | −0.102 | 0.370 | −0.002 | 0.984 | 0.013 | 0.914 | |||
| AST | 0.118 | 0.076 | −0.068 | 0.553 | −0.023 | 0.844 | 0.167 | 0.165 | |||
| ALT | 0.113 | 0.089 | −0.056 | 0.625 | 0.106 | 0.359 | 0.108 | 0.369 | |||
NGT, normal glucose tolerance; T2DM, type 2 diabetes mellitus; BMI, body mass index; BP, blood pressure; Hb, hemoglobin; HbA1c, glycosylated hemoglobin; PP2, 2 hour postprandial; HOMA-IR, homeostasis model assessment of insulin resistance; HOMA-β, homeostasis model assessment of β-cell function; TG, triglyceride; TC, total cholesterol; HDL-C, high density lipoprotein cholesterol; LDL-C, low density lipoprotein cholesterol; BUN, blood urea nitrogen; eGFR, Estimated glomerular filtration rate; AST, Aspartate aminotransferase; ALT, Alanine aminotransferase.
Estimated sLRIG2 level after adjusting for various covariates
Factors that significantly affected the difference in LRIG2 level among the three groups were investigated. In addition, serum sLRIG2 levels were compared among the three groups after adjusting for several clinical factors. After adjusting for the various factors, except glucose parameters, a significant difference was observed among the three groups, similar to before correction (all P<0.01). However, the significant difference among the three groups disappeared after adjustment for HbA1c (P=0.258) and glucose (fasting: P=0.203, 2-hour postprandial: P=0.700; Table 3).
Table 3
| Covariate | Estimated sLRIG2 value ± SE (95% CI) | P value (post-hoc) | ||
|---|---|---|---|---|
| NGT | Prediabetes | T2DM | ||
| Height | 12.4±0.9 (10.6–14.3) | 13.4±0.9 (11.6–15.3) | 16.7±0.8 (15.1–18.2) | 0.001 (T2DM > NGT, Prediabetes) |
| Weight | 12.6±0.9 (10.7–14.4) | 13.5±0.9 (11.7–15.4) | 16.7±0.8 (15.1–18.2) | 0.002 (T2DM > NGT, Prediabetes) |
| Diastolic BP | 12.5±0.9 (10.7–14.3) | 13.8±0.9 (12.1–15.6) | 16.8±0.8 (15.2–18.3) | 0.001 (T2DM > NGT, Prediabetes) |
| HbA1c | 13.2±0.8 (11.6–14.7) | 13.6±0.8 (12.2–15.2) | 15.3±0.9 (13.6–17.1) | 0.258 |
| Glucose, fasting | 13.2±0.8 (11.7–14.7) | 13.6±0.7 (12.1–15.0) | 15.4±0.9 (13.7–17.1) | 0.203 |
| Glucose, PP2 | 13.6±0.9 (11.9–15.3) | 13.7±0.8 (12.2–15.2) | 14.9±1.0 (12.8–16.9) | 0.700 |
| Insulin, fasting | 12.1±0.7 (10.7–13.6) | 12.9±0.7 (11.4–14.3) | 16.9±0.8 (15.5–18.4) | <0.001 (T2DM > NGT, Prediabetes) |
| C-peptide, fasting | 11.9±0.8 (10.3–13.4) | 13.0±0.8 (11.4–14.5) | 16.5±0.8 (15.0–18.1) | <0.001 (T2DM > NGT, Prediabetes) |
| HOMA-IR | 12.4±0.8 (10.9–13.9) | 13.0±0.7 (11.5–14.5) | 16.5±0.8 (14.9–18.1) | 0.001 (T2DM > NGT, Prediabetes) |
| HOMA-β | 12.2±0.7 (10.8–13.6) | 13.0±0.7 (11.5–14.4) | 16.8±0.7 (15.4–18.2) | <0.001 (T2DM > NGT, Prediabetes) |
| TG | 12.5±0.7 (11.1–14.0) | 13.0±0.7 (11.6–14.5) | 16.7±0.8 (15.2–18.2) | <0.001 (T2DM > NGT, Prediabetes) |
| HDL-C | 12.4±0.8 (10.9–13.9) | 13.0±0.7 (11.5–14.5) | 16.8±0.8 (15.3–18.4) | <0.001 (T2DM > NGT, Prediabetes) |
P values obtained from analyses of covariance using the post hoc Bonferroni correction. NGT, normal glucose tolerance; T2DM, type 2 diabetes mellitus; BP, blood pressure; HbA1c, glycosylated hemoglobin; PP2 glucose, 2-hour postprandial glucose; HOMA-IR, homeostasis model assessment of insulin resistance; HOMA-β, homeostasis model assessment of β-cell function; TG, triglyceride; HDL-C, high density lipoprotein cholesterol; CI, confidential interval.
Diagnostic performance of serum sLRIG2 in diagnosis of type 2 diabetes
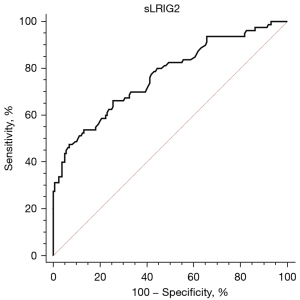
Diagnostic ability and optimal cutoff value of serum sLRIG2 for the diagnosis type 2 diabetes were assessed by evaluating sensitivity and specificity using ROC curve. The area under the ROC curve was 0.768 and the best cut-off value for serum sLRIG2 was 15.1 ng/mL (sensitivity 66.25%, specificity 74.37%) (Figure 3).
Discussion
As type 2 diabetes is a growing worldwide health problem, there is a need to develop effective diagnostic biomarker for diabetes. Numbers of molecules, such as high-sensitivity C-reactive protein (hsCRP), fibroblast growth factor 21 (FGF21), adiponectin and microRNA, have been proposed to be potential biomarker (21-24). However, these biomarkers have limitations in the early detection of diabetes. To investigate the usefulness of serum soluble LRIG2 as biomarker for the early diagnosis of type 2 diabetes, we enrolled only subject with drug-naïve, newly diagnosed type 2 diabetes. The results confirmed that sLRIG2 levels are significantly higher in subjects with glucose intolerance. We demonstrated the possible utility of sLRIG2 as a potential diagnostic biomarker of type 2 diabetes.
LRIG2, a member of the LRIG gene family, is a regulator of growth factor receptor (4). Similar to other members of the LRIG family, LRIG2 has been mainly considered in cancer research. In previous studies, increased LRIG2 expression was reported in various cancers with increased EGFR expression. Downregulation of LRIG2 expression resulted in decreased EGFR expression, inhibition of cell growth, and increased apoptosis (25,26). The LRIG gene family encodes integral membrane proteins consisting of an ectodomain, a transmembrane domain, and a cytoplasmic tail (27). In several studies, all LRIG proteins were shown to have a soluble form. The soluble LRIG ectodomain appears to have a function similar to their corresponding full-length protein (10,28). The function of soluble forms in the LRIG family has been examined in a few studies; however, the role of sLRIG2 was reported in only one study, by Xiao at al. The authors demonstrated that sLRIG2 ectodomain is released from glioblastoma cells. Furthermore, the physical interaction of the LRIG2 ectodomain with EGFR and sLRIG2 reportedly has potent pro-tumor effects similar to the full-length form of LRIG2 (27). In addition to cancers, urofacial syndrome was shown to be associated with LRIG2 mutation (29). However, the roles of LRIG2 and sLRIG2 in metabolic disorders have not been examined. This is the first study in which the association between metabolic disorder and sLRIG2 was investigated.
We previously confirmed that serum sEGFR levels are higher in subjects with newly diagnosed type 2 diabetes compared with controls (18). Because both sEGFR and sLRIG2 are elevated in type 2 diabetes compared with controls, the mechanism linking sLRIG2 levels with glucose intolerance may involve EGFR. The association between LRIG2 and glucose metabolism has not been examined in previous research; however, a role of EGFR signaling in metabolic regulation has been established. EGFR signaling enhances aerobic glycolysis in cancer cells (30,31). Furthermore, inhibition of EGFR signaling was shown to improve insulin sensitivity in several studies (14,32). Reportedly, treatment with the specific EGFR inhibitor, PD153035, reduced fasting glucose and fasting insulin, and improved glucose intolerance and insulin resistance in high-fat diet (HFD)-fed mice (15). However, in a recent study, an EGFR-independent role of LRIG was discovered. Herdenberg et al. suggested that the LRIG1 protein is a regulator of bone morphogenetic protein (BMP) signaling, and LRIG1 gene variants were associated with a decreased risk of type 2 diabetes (19). The possibility that LRIG2 participates in metabolic regulation in a different manner than a receptor tyrosine kinase regulator cannot be excluded.
In the present study, among parameters of glucose homeostasis, positive correlations were found between serum sLRIG2 levels and HbA1c, fasting plasma glucose, and 2-hour postprandial blood glucose. However, no significant correlations were observed between sLRIG2 levels and glucose parameters in subjects with NGT or prediabetes. In addition, sLRIG2 and HOMA-IR were positively correlated in all subjects. This indicates that sLIRG2 is a potential biomarker for diabetes and an indicator of insulin resistance in patients newly diagnosed with type 2 diabetes. In addition to glucose parameters, body weight was significantly correlated with sLRIG2 levels. Herdenberg et al. demonstrated reduced adipocyte differentiation in LRIG null mouse embryonic fibroblasts (MEFs) (19). LRIG2 expression levels were also found to be upregulated during the adipogenic process, indicating that LRIG2 may contribute to adipogenesis. In the present study, the sLRIG2 level was significantly different among the three groups. To determine whether various clinical factors, including body weight, affect the sLRIG2 levels, those factors were adjusted for. After adjusting for glucose parameters, the significant difference that existed before the correction disappeared; however, changes in other factors, such as body weight, were not observed after adjustment; thus, clinical factors other than glucose parameters were not expected to have a significant mediating effect on the difference in sLRIG2 levels among the three groups.
The present study had several limitations. Because this was a retrospective and cross-sectional study, whether changes in glucose and HbA1c actually induce changes in sLRIG2 could not be investigated. And we also couldn’t determin whether the acute glucose fluctuation affetc serum LRIG2 levels because we measured sLRIG2 levels from a fasting sample only excluding postprandial sample. In addition, a molecular mechanism linking LRIG2 with glucose metabolism was not confirmed. Further research is needed to determine whether the molecular mechanism is EGFR-dependent or -independent.
In conclusion, serum sLRIG2 levels were confirmed to be higher in subjects with type 2 diabetes. Furthermore, serum sLRIG2 levels showed significant correlations with plasma glucose and HbA1c, and the usefulness of serum sLRIG2 level as a potential novel biomarker of type 2 diabetes was demonstrated.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-3272
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-3272
Peer Review File: Available at https://dx.doi.org/10.21037/atm-21-3272
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-3272). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the Institutional Review Board of Chungnam National University Hospital (CNUH 2014-12-013) and written informed consent was obtained from each participant before enrollment.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Saeedi P, Petersohn I, Salpea P, et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res Clin Pract 2019;157:107843.
- Plantinga LC, Crews DC, Coresh J, et al. Prevalence of chronic kidney disease in US adults with undiagnosed diabetes or prediabetes. Clin J Am Soc Nephrol 2010;5:673-82. [Crossref] [PubMed]
- Gossain VV, Aldasouqi S. The challenge of undiagnosed pre-diabetes, diabetes and associated cardiovascular disease. Int J Diabetes Mellit 2010;2:43-6. [Crossref]
- Simion C, Cedano-Prieto ME, Sweeney C. The LRIG family: enigmatic regulators of growth factor receptor signaling. Endocr Relat Cancer 2014;21:R431-43. [Crossref] [PubMed]
- Powell AE, Wang Y, Li Y, et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 2012;149:146-58. [Crossref] [PubMed]
- Thomasson M, Hedman H, Guo D, et al. LRIG1 and epidermal growth factor receptor in renal cell carcinoma: a quantitative RT--PCR and immunohistochemical analysis. Br J Cancer 2003;89:1285-9. [Crossref] [PubMed]
- Hedman H, Lindström AK, Tot T, et al. LRIG2 in contrast to LRIG1 predicts poor survival in early-stage squamous cell carcinoma of the uterine cervix. Acta Oncol 2010;49:812-5. [Crossref] [PubMed]
- Holmlund C, Haapasalo H, Yi W, et al. Cytoplasmic LRIG2 expression is associated with poor oligodendroglioma patient survival. Neuropathology 2009;29:242-7. [Crossref] [PubMed]
- Rafidi H, Mercado F 3rd, Astudillo M, et al. Leucine-rich repeat and immunoglobulin domain-containing protein-1 (Lrig1) negative regulatory action toward ErbB receptor tyrosine kinases is opposed by leucine-rich repeat and immunoglobulin domain-containing protein 3 (Lrig3). J Biol Chem 2013;288:21593-605. [Crossref] [PubMed]
- Cheng F, Zhang P, Xiao Q, et al. The Prognostic and Therapeutic Potential of LRIG3 and Soluble LRIG3 in Glioblastoma. Front Oncol 2019;9:447. [Crossref] [PubMed]
- Guo D, Yang H, Guo Y, et al. LRIG3 modulates proliferation, apoptosis and invasion of glioblastoma cells as a potent tumor suppressor. J Neurol Sci 2015;350:61-8. [Crossref] [PubMed]
- Normanno N, De Luca A, Bianco C, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006;366:2-16. [Crossref] [PubMed]
- Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. FEBS J 2010;277:301-8. [Crossref] [PubMed]
- Li Z, Li Y, Overstreet JM, et al. Inhibition of Epidermal Growth Factor Receptor Activation Is Associated With Improved Diabetic Nephropathy and Insulin Resistance in Type 2 Diabetes. Diabetes 2018;67:1847-57. [Crossref] [PubMed]
- Choung S, Kim JM, Joung KH, et al. Epidermal growth factor receptor inhibition attenuates non-alcoholic fatty liver disease in diet-induced obese mice. PLoS One 2019;14:e0210828 [Crossref] [PubMed]
- Miettinen P, Ormio P, Hakonen E, et al. EGF receptor in pancreatic beta-cell mass regulation. Biochem Soc Trans 2008;36:280-5. [Crossref] [PubMed]
- Miettinen PJ, Ustinov J, Ormio P, et al. Downregulation of EGF receptor signaling in pancreatic islets causes diabetes due to impaired postnatal beta-cell growth. Diabetes 2006;55:3299-308. [Crossref] [PubMed]
- Kim JM, Choung S, Joung KH, et al. Serum Soluble Epidermal Growth Factor Receptor Level Increase in Patients Newly Diagnosed with Type 2 Diabetes Mellitus. Diabetes Metab J 2018;42:343-7. [Crossref] [PubMed]
- Herdenberg C, Mutie PM, Billing O, et al. LRIG proteins regulate lipid metabolism via BMP signaling and affect the risk of type 2 diabetes. Commun Biol 2021;4:90. [Crossref] [PubMed]
- Matthews DR, Hosker JP, Rudenski AS, et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412-9. [Crossref] [PubMed]
- Vinagre I, Sánchez-Quesada JL, Sánchez-Hernández J, et al. Inflammatory biomarkers in type 2 diabetic patients: effect of glycemic control and impact of LDL subfraction phenotype. Cardiovasc Diabetol 2014;13:34. [Crossref] [PubMed]
- Tabák AG, Carstensen M, Witte DR, et al. Adiponectin trajectories before type 2 diabetes diagnosis: Whitehall II study. Diabetes Care 2012;35:2540-7. [Crossref] [PubMed]
- Al-Kafaji G, Al-Mahroos G, Alsayed NA, et al. Peripheral blood microRNA-15a is a potential biomarker for type 2 diabetes mellitus and pre-diabetes. Mol Med Rep 2015;12:7485-90. [Crossref] [PubMed]
- Woo YC, Lee CH, Fong CH, et al. Serum fibroblast growth factor 21 is a superior biomarker to other adipokines in predicting incident diabetes. Clin Endocrinol (Oxf) 2017;86:37-43. [Crossref] [PubMed]
- Yang HK, Chen H, Mao F, et al. Downregulation of LRIG2 expression inhibits angiogenesis of glioma via EGFR/VEGF-A pathway. Oncol Lett 2017;14:4021-8. [Crossref] [PubMed]
- Wang B, Han L, Chen R, et al. Downregulation of LRIG2 expression by RNA interference inhibits glioblastoma cell (GL15) growth, causes cell cycle redistribution, increases cell apoptosis and enhances cell adhesion and invasion in vitro. Cancer Biol Ther 2009;8:1018-23. [Crossref] [PubMed]
- Xiao Q, Tan Y, Guo Y, et al. Soluble LRIG2 ectodomain is released from glioblastoma cells and promotes the proliferation and inhibits the apoptosis of glioblastoma cells in vitro and in vivo in a similar manner to the full-length LRIG2. PLoS One 2014;9:e111419 [Crossref] [PubMed]
- Goldoni S, Iozzo RA, Kay P, et al. A soluble ectodomain of LRIG1 inhibits cancer cell growth by attenuating basal and ligand-dependent EGFR activity. Oncogene 2007;26:368-81. [Crossref] [PubMed]
- Roberts NA, Hilton EN, Lopes FM, et al. Lrig2 and Hpse2, mutated in urofacial syndrome, pattern nerves in the urinary bladder. Kidney Int 2019;95:1138-52. [Crossref] [PubMed]
- Lim SO, Li CW, Xia W, et al. EGFR Signaling Enhances Aerobic Glycolysis in Triple-Negative Breast Cancer Cells to Promote Tumor Growth and Immune Escape. Cancer Res 2016;76:1284-96. [Crossref] [PubMed]
- Makinoshima H, Takita M, Matsumoto S, et al. Epidermal growth factor receptor (EGFR) signaling regulates global metabolic pathways in EGFR-mutated lung adenocarcinoma. J Biol Chem 2014;289:20813-23. [Crossref] [PubMed]
- Zhao M, Jung Y, Jiang Z, et al. Regulation of Energy Metabolism by Receptor Tyrosine Kinase Ligands. Front Physiol 2020;11:354. [Crossref] [PubMed]


