
Klotho upregulates the interaction between RANK and TRAF6 to facilitate RANKL-induced osteoclastogenesis via the NF-κB signaling pathway
Introduction
Bone is a dynamic tissue that is continuously remodeled by osteoblasts and osteoclasts. Bone integrity and mineral homeostasis rely on a balance between osteoblastic bone formation and osteoclastic bone resorption (1). Osteoclasts are derived from hematopoietic stem cells, and they are the only cell type responsible for bone resorption in adulthood. Abnormalities of osteoclasts result in many diseases such as osteoporosis, ectopic ossification, and osteopetrosis (2). Studies on osteoclastogenesis can provide valuable strategies for combating osteoclast abnormalities.
Klotho (KL) is a highly conserved gene that encodes the klotho protein, with a sequence homology of up to 94% among humans, rats, and mice (3-5). Importantly, klotho mutant (kl/kl) mice that carry hypomorph klotho alleles develop a syndrome exhibiting human aging-related phenotypes, including short life span (3–4 months), neural degeneration, and abnormal mineral metabolism. An interesting finding in kl/kl mice is that both osteoblasts and osteoclasts are impaired and lead to low-turnover osteoporosis, however, this impairment is independent of Osteoblast-Osteoclast Interactions. Osteoclast abnormality has no relationship with a defect in osteoblastic-cell support of osteoclast differentiation but is due to abnormalities of osteoclast progenitors (6). It is difficult to conclude that klotho solely leads to the abnormality of osteoclast progenitors in kl/kl mice as klotho is completely deficient in this model. Also, the role of klotho during osteoclastogenesis remains unclear, so it is of great value to explore the function of klotho in osteoclastogenesis and the underlying mechanisms.
Osteoclastogenesis relies on the stimulation of 2 important factors, namely macrophage colony-stimulating factor (M-CSF) and receptor activator of nuclear factor kappa-B ligand (RANKL) (7,8). M-CSF is a cytokine involved in the initiation of bone marrow-derived macrophages (BMMs) differentiation into osteoclast precursors, and it also regulates the survival and proliferation of BMMs and pre-osteoclasts. RANKL is the only ligand that binds to the extracellular portion of RANK (9). It induces osteoclast differentiation, the termination of differentiation, and supports the differentiation of pre-osteoclasts into mature osteoclasts (10). RANK, a type I transmembrane protein, recruits different adaptor proteins for intracellular signal transduction, followed by the activation of various signaling pathways such as NF-κB, ERK, p38, and AKT (1,2,11-14). Among these adaptor proteins, TNFR-associated factors (TRAFs), especially TRAF6, are the most critical ones. TRAF6 can be recruited by RANK after RANKL stimulation to form a RANK-TRAF6 complex through which downstream signaling pathways are activated (15,16). Various activated downstream signaling pathways ultimately trigger the expression and translocation of nuclear factor of activated T cells cytoplasmic 1 (NFATc1), which serves as an essential regulator for a number of osteoclast-specific genes responsible for osteoclast function such as TRAP, cathepsin K (CTSK), and calcitonin receptor (CTR) through cooperation with c-Fos (17-19).
In this study, we used kl/kl mice and several in vitro experiments which demonstrated that klotho promotes RANKL-induced osteoclastogenesis. Through an exploration of the underlying mechanisms, during RANKL-induced Osteoclastogenesis, klotho activates the downstream NF-κB signaling pathway thereby upregulating the activation of the expression of c-Fos and NFATc1, moreover, we found that the positive effect of klotho on the activation of NF-κB signaling pathway was attributed to klotho binds to RANK to facilitate the interaction between RANK and TRAF6. Klotho may serve as a promising therapeutic target for osteoclasts related diseases. We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-4332).
Methods
Animals
We purchased heterozygous klotho mutant mice (kl/+) named C57BL/6N-Klem1cyagen from Cyagen Biosciences Inc. Since homozygous klotho mutant mice (kl/kl) are infertile, we generated kl/kl mice and wild-type mice (WT) by crossing kl/+ mice. All the experimental mice were 4–6 weeks old. Klotho mutant homozygotes or WT mice (4–6 weeks old) were used for experiments. The kl/kl mice and WT mice were bred and maintained in the animal facilities at the Third Military Medical University. Experiments were performed under a project license (NO.: AMUWEC2021881) granted by the ethics board of The Third Military Medical University, in compliance with The Third Military Medical University institutional guidelines for the care and use of animals.
MicroCT analysis
The mice were sacrificed and We dissected femur specimens of both sides from kl/kl mice and WT mice, all adapted mice were 4-week-old males. Then specimens were fixed overnight in 10% formalin and analyzed by high-resolution µCT (Skyscan1272, Bruker microCT, Kontich, Belgium). There were 24 samples in total, including 12 specimens from kl/kl mice and 12 specimens from WT mice. The scanner was set at a voltage of 60 kV and a resolution of 12 µm per pixel. Images of perfusion computed tomography (PCT) were used to perform three-dimensional (3D) histomorphometric analyses. The region of interest was defined to cover the whole PCT compartment (the femoral head and the femoral shaft). The images were reconstructed with NRecon v1.6 software (Bioz, Inc., CA, USA), analyzed by CTAn v1.9 software (Bruker microCT), and visualized using the 3D model visualization software CTVol v2.0 (Bruker microCT).
Histochemistry
We used femur specimens from 4-week-old males kl/kl mice and WT mice for histochemistry analysis. After 4 weeks bone decalcification in 4 °C, specimens were embedded as paraffin blocks. Blocks were sectioned at 4 µm using a paraffin microtome. We performed staining using paraffin-embedded sections. The sections were dewaxed and then washed 3 times with phosphate-buffered saline (PBS). The sections were used for tartrate-resistant acid phosphatase (TRAP) staining, which was conducted in accordance with the protocol provided by the manufacturer (387A-1KT, Sigma-Aldrich, USA), followed by counterstaining with methyl green (M884, Sigma-Aldrich, MO, USA) for 10 s. A total of 12 blocks were used for analysis, including 6 femurs from kl/kl mice and 6 femurs from WT mice.
Isolation of BMMs
BMMs were isolated from the femurs and tibias of kl/kl mice and WT mice, and all adapted mice were 4-week-old males. The mice were sacrificed and the hind legs were sterilized with 70% ethanol. Followed by removing all the connected soft tissues from the bones and their femurs and tibias were dissected. After the epiphysis is removed, the bone marrow was flushed using alpha-modified minimal essential medium (α-MEM) (HyClone, UT, USA) and the cells in medium were considered as bone marrow cells. The bone marrow cells were mixed with red blood cell lysis buffer (Beyotime, Shanghai, China) for 2 min, then the cells were cultured for 12 h. The non-adherent cells were collected and the concentration was adjusted to 2×106 cells/mL in BMM medium (α-MEM) containing 10% fetal bovine serum (FBS, HyClone, UT, USA) and 30 ng/mL M-CSF (R&D Systems, MN, USA), then cells were placed in a humidified incubator with 5% CO2 at 37 °C. After 2 days without any procedures, the culture medium was removed and the cells that adhered to the dish were considered BMMs which could be used for subsequent experiments.
Cell culture and osteoclast differentiation
RAW264.7 cells were obtained from Shanghai Institutes for Biological Sciences (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle medium (DMEM) (HyClone, UT, USA) containing 10% FBS (HyClone, UT, USA). For osteoclast differentiation using RAW264.7 cells, the cells were pretreated with M-CSF (30 ng/mL) for 1d followed by exposure to RANKL (50 ng/mL) (R&D Systems, Minneapolis, MN, USA) to stimulate osteoclastogenesis. For osteoclastogenesis using BMMs obtained as described above, cells were cultured in α-MEM containing 10% FBS, M-CSF (30 ng/mL), and RANKL (50 ng/mL). Fixed cells were stained with TRAP using the TRAP staining kit (Sigma, USA) according to the manufacturer’s instructions. TRAP+ multinucleated cells, including more than 3 nuclei, were counted. Quantifications were performed using the software ImageJ 1.53c (National Institutes of Health, MD, USA). In some experiments, 2.5 µM of BAY 11-7082 (Beyotime, Shanghai, China) was included. Culture media was replaced every 2 days.
Lentivirus transfection
LV-NCKL, LV-KL, LV-NCshKL, and LV-shKL were supplied by Hanbio (Shanghai, China). The lentiviruses were purified from supernatants using ultracentrifugation (3,000 g and 4 °C for 15 min) 72 h post-transfection, and the titers of the lentiviruses were determined, we adapted MOI 50 for transfection. RAW264.7 cells were transfected for 8 h and 3 times each, then puromycin (3 µg/mL) was used for selection. Cells which expressed GFP were considered as transfected cells and were used to measure transfection efficiency.
Immunohistochemistry
RANKL-induced RAW264.7 cells were seeded in 96-well plates at a starting density of 2×104 cells/well. On 3 d, cells were washed using PBS 3 times and fixed with 3.75% formaldehyde in cold PBS for 10 min, followed by 0.5% Triton X-100 for 1-min permeabilization. Blocking was then performed using 5% skim milk overnight at 4 °C, and cells were incubated in primary antibody solution for 1 h at room temperature using reagents from the Actin Cytoskeleton and Focal Adhesion Staining kit (Merck Millipore, Darmstadt, Germany). Cells were then washed with PBS 3 times and incubated with the secondary antibody for 1 h at room temperature. Cells were counterstained with DAPI for 10 min followed by observation with fluorescence microscopy.
Pit formation
BMMs isolated from the femurs and tibias of either kl/kl mice or WT mice as previous described were used for pit formation, Bovine bone slices (IDS Nordic, Herlev, Denmark) were placed in 48-well plates and 4×104 BMMs were seeded per well. Cells were stimulated with M-CSF (50 ng/mL) and RANKL (50 ng/mL) to generate multinucleated osteoclasts for 5 d, then the bone slices were washed with PBS 3 times. Then, 1N NaOH was used to remove the adherent cells, and resorption pits were visualized by staining with hematoxylin (Beyotime, Shanghai, China) for 3 mins. The pit formation area was analyzed by ImageJ 1.53c software (National Institutes of Health, MD, USA).
RT-qPCR
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Single-stranded cDNA was reverse transcribed from 1 µg of total RNA according to the manufacturer’s instructions of Reverse Transcription System kit (Promega). Quantitative PCR was performed on CFX96 Touch Real-Time PCR System (Bio-Rad, CA, USA) using Power SYBR-Green PCR Master Mix (Takara, Shiga, Japan) according to the instructions. The specific primer sequences were designed as follows: 5'-AAAGCTGATAGAGGACAATGGC-3' (Klotho, forward), 5'-GGCAGAGAAATCAACACAGTAAGG-3' (Klotho, reverse); 5'-GACCCGGAGTTCGACTTCG-3' (NFATc1, forward), 5'-TGACACTAGGGGACACATAACTG-3' (NFATc1, reverse); 5'-CGGGTTTCAACGCCGACTA-3' (c-Fos, forward), 5'-TTGGCACTAGAGACGGACAGA-3' (c-Fos, reverse); 5'-GAAGAAGACTCACCAGAAGCAG-3' (CTSK, forward), 5'-TCCAGGTTATGGGCAGAGATT-3′ (CTSK, reverse); 5'-CGCATCCGCTTGAATGTG-3' (CTR, forward), 5'-TCTGTCTTTCCCCAGGAAATGA-3' (CTR, reverse); 5'-TGTATCGGCTCATCTCCTCCAT-3' (DC-STAMP, forward), 5'-GACTCCTTGGGTTCCTTGCTT-3' (DC-STAMP, reverse); 5'-AGATGTGGATCAGCAAGCAG-3' (β-actin, forward), 5'-GCGCAAGTTAGGTTTTGTCA-3' (β-actin, reverse). β-actin served as the internal housekeeping gene control.
Western blot
Cells were lysed in cell lysis buffer for western blot and Co-IP (Beyotime, Shanghai, China) and 25 µg of protein from each sample was subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). After transferring the proteins onto PVDF membranes (0.22 µm; Merck Millipore, Darmstadt, Germany), followed by blocking in 5% skim milk for 2 h, the membranes were incubated with the following primary antibodies overnight at 4 °C: klotho (1:500; Bioss, Beijing, China), NFATc1 (1:1,000; Abcam, Cambridge, UK), c-Fos (1:500; Bioss, Beijing, China), and GAPDH (1:1,000; Bioss, Beijing, China). Then, the membranes were washed 3 times with Tris-buffered saline and tween (TBST) followed by incubation with anti-rabbit secondary antibodies (1:1,000; Bioss, Beijing, China) in a blocking solution for 1 h at room temperature. Blots against GAPDH were set as the loading controls. For the detection of NF-κB signaling pathway phosphorylation, IκB, P65, and their phosphorylated proteins were detected. The medium of RANKL-induced BMMs and RAW 264.7 cells at 1 d was replaced by medium without RANKL for 12 h, then cells were treated with or without RANKL for 10 min. The subsequent steps for western blot were the same as mentioned above. Primary antibodies against IκB (1:500), p-IκB (1:500), P65 (1:1,000), and p-P65 (1:1,000) were all supplied by Abcam (Cambridge, UK).
RNA-seq analysis and differential pathway analysis
Total RNA was extracted using TRIzol from RANKL-induced BMMs of either kl/kl mice or WT mice at 1 d, and ribosomal RNA was removed using the Ribo-Zero™ kit (Epicentre, Madison, WI, USA). The purified library products were prepared according to the protocol of the NEBNext®Ultra™ RNA Library Prep kit for Illumina (NEB, MA, USA), then evaluated with the Agilent 2200 TapeStation and Qubit®2.0 (Life Technologies, MD, USA). The libraries were sequenced with the IlluminaHiSeq 3000 platform at Guangzhou RiboBio Co. Ltd. (Guangzhou, China). The limma R package was used for gene expression analysis, then Gene Ontology (GO) analysis and KEGG pathway enrichment were performed using the cluster Profiler R package.
Energy-based protein docking analysis
The protein structure of RANK (ID: 3ME2) was downloaded from the Protein Data Bank (PDB) database. The 3D protein model of the extracellular region of α-klotho (NP_038851.2) was constructed using the Rosetta 49–51 program (20-22). Rosetta was used to predict an automated 3D structure of klotho, where the benchmarked scoring system helps to obtain quantitative assessments of the Rosetta models. To select the final models, Rosetta clustered all the decoys based on the pairwise structure similarity and reported up to 5 models which corresponded to the 5 largest structure clusters. The top-scoring model was used for future analysis. The ZDOCK program was primarily used to search for all possible modes of interaction in the space between 2 proteins by translation and rotation, and to evaluate each binding model using an energy-based scoring function (23). Docking simulations were run with ZDOCK to generate rigid-body docking poses, which were rescored by energy-based functions composed of van der Waals, electrostatics, and solvation energy terms (24).
Co-IP assay
RAW264.7 cells were treated with RANKL, then the supernatant was obtained after homogenization and centrifugation, followed by incubating with beads bound to the beads bound to the anti-RANK antibody. Centrifugation of the mixture was performed and the supernatant was discarded. The beads were washed and the protein complex was collected. The western blot assay as described above was used for detection. Anti-GAPDH antibody (Bioss, Beijing, China) was used as the loading control.
Statistical analysis
The data were statistically analyzed using Statistical Product and Service Solutions (SPSS) version 15 software (IBM, Armonk, NY, USA), and were presented as mean ± SD. An unpaired two-tailed Student’s t-test was conducted for comparisons between 2 groups. The level of significance was set at P<0.05, GraphPad Prism 8 software was employed for statistical analysis.
Results
Deficiency of klotho impairs osteoclastogenesis
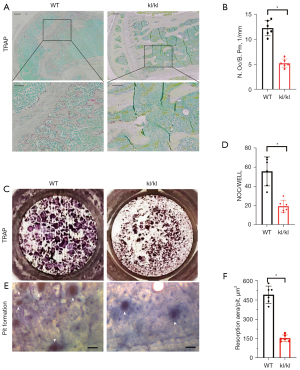
We collected femur specimens of kl/kl mice and WT mice which were then systemically scanned and analyzed by micro-computed tomography (µCT). The images showed that there was increased trabecular bone in the kl/kl group compared with the sham group, whereas cortical bone decreased (Figure S1A,S1B). Impaired osteoclastogenesis in kl/kl mice was indicated by significantly lower values of Oc.s/BS (%) and N.OC/B.Pm compared with WT mice (Figure S1B). A previous study identified that klotho causes impairment of osteoclasts independently from osteoblasts (3), but its effect on osteoclastogenesis remains unknown. Therefore, tartrate-resistant acid phosphatase (TRAP) staining of femur heads was performed and revealed that the number of TRAP + osteoclasts was significantly decreased in kl/kl mice compared to WT mice (Figure 1A,1B). To further assess the effects of klotho deficiency on osteoclast formation and function, a RANKL-stimulated osteoclastogenesis assay was performed in BMMs which were isolated from kl/kl mice and WT mice separately. As shown by the results, there were dramatically fewer TRAP + multinucleated cells of the kl/kl group compared to the sham controls (Figure 1C,1D). The pit formation assay was carried out to evaluate the effects of klotho deficiency on osteoclastic bone resorption using bovine bone slices, which showed that the bone resorption of osteoclasts significantly decreased in the kl/kl group compared with the WT group, suggesting reduced osteoclast function (Figure 1E,1F). Taken together, we concluded that deficiency of klotho impaired osteoclastogenesis.
Klotho promotes osteoclastogenesis in vitro
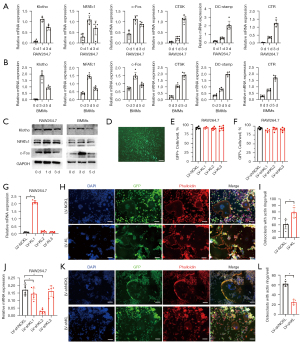
To examine the role of klotho during osteoclastogenesis, RT-qPCR was employed in both RANKL-treated RAW264.7 cells and BMMs from WT mice to detect the transcriptional levels of klotho, c-Fos, NFATc1, and several other osteoclast differentiation-related marker genes (CTSK, CTR, DC-stamp). The results revealed that klotho was expressed in osteoclastic cells and shared the same expression pattern with NFATc1 and c-Fos during osteoclastogenesis, which reached their expression peak at the early phase of osteoclast differentiation, unlike the other related marker genes (CTSK, CTR, DC-stamp) (Figure 2A,2B). Furthermore, the measurement of protein expression by western blot also showed consistent results (Figure 2C). It has been well established that NFATc1 in cooperation with c-Fos are master regulators of osteoclastogenesis marker genes (25-27). The above results suggested that klotho might exert functions during osteoclastogenesis. To confirm this, we transfected RAW264.7 cells using a lentivirus with a GFP expression vector for stable overexpression or knockdown of klotho in cells. Transfection efficiency was tested by GFP+ cells/well after puromycin selection (Figure 2D-2F), and the transfection effect was tested by RT-qPCR for klotho mRNA level. Three experimental vectors for each group were tested, and the subgroup LV-KL1 with the highest transfection effect were chosen for the subsequent experiments (Figure 2G). We found that wells with klotho overexpression in RANKL-treated RAW264.7 cells had more osteoclasts with actin rings compared to the sham controls at 3 d (Figure 2H,2I), and under the same condition, the subgroup LV-shKL2 with the highest transfection effect were chosen for the subsequent experiments (Figure 2J). Klotho knockdown in RAW264.7 cells led to decreased osteoclasts with actin rings (Figure 2K,2L). Collectively, these results revealed that klotho facilitates RANKL-induced osteoclastogenesis in vitro.
Klotho upregulates the expression of NFATc1 and c-Fos during osteoclastogenesis
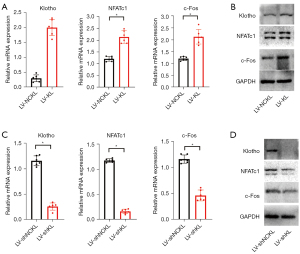
To further explore whether klotho affects essential regulators during osteoclastogenesis, we carried out RT-qPCR and western blot and found that overexpression of klotho in RANKL-treated RAW264.7 cells increased the expression of NFATc1 and c-Fos at the mRNA level and protein level at 1 d compared to sham controls (Figure 3A,3B). Knockdown of klotho in RANKL-treated RAW264.7 cells decreased the expression of NFATc1 and c-Fos at the mRNA level and protein level at 1d compared to sham controls (Figure 3C,3D). These results suggested that klotho positively regulates the expression of NFATc1 and c-Fos during osteoclastogenesis.
Klotho exerts functions through the NF-κB signaling pathway
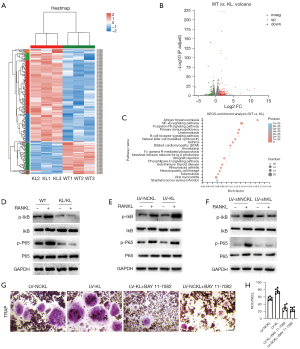
To explore the underlying mechanisms by which klotho promotes RANKL-induced osteoclastogenesis, we firstly identified differentially expressed mRNAs in kl/kl mice and WT mice which were presented in a heatmap and volcano plots (Figure 4A,4B). A total of 2,335 differentially expressed mRNAs were identified between kl/kl mice and WT mice. Of these, 1,120 were upregulated and 1,215 were downregulated in kl/kl mice. Then, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was used to examine the processes in which the differentially expressed mRNAs were involved. The results revealed that the ‘NF-κB signaling pathway’ was the most enriched pathway (Figure 4C), suggesting that this was the primary signaling pathway in which differentially expressed mRNAs were involved. According to the above results, we employed western blot to detect whether klotho exerted any promotive impact on the RANKL-induced phosphorylation of specific downstream signals of the NF-κB pathway. The results showed that the phosphorylation of IκB and P65 by RANKL was not significantly different in BMMs isolated from kl/kl mice compared with the elevated phosphorylation levels of IκB and P65 in BMMs from WT mice (Figure 4D). In addition, the elevated phosphorylation levels of IκB and P65 by RANKL were more significant in klotho overexpressed RAW264.7 cells compared with sham controls (Figure 4E). However, these 2 factors showed no significant changes in klotho knockdown RAW264.7 cells compared with sham controls (Figure 4F). Furthermore, the effect of klotho on osteoclastogenesis in RANKL-treated RAW264.7 cells was significantly suppressed after treatment with BAY 11-7082, which is a widely used inhibitor of the NF-κB pathway (Figure 4G,4H). Taken together, the above results suggested that klotho facilitates osteoclastogenesis by upregulating the activation of the NF-κB pathway.
Klotho combines with RANK to promote the interaction between RANK and TRAF6
To further identify the underlying mechanism of NF-κB pathway activation, RT-qPCR and western blot were performed to explore the changes upstream of the NF-κB pathway. To our surprise, mRNA and protein levels of RANK and TRAF6 in RANKL-treated RAW264.7 cells at 1d showed no statistically significant differences between either the LV-KL group or the LV-shLV group compared to sham controls (Figure 5A-5C). These data suggested that klotho might exert its function through mediating the interaction between RANK and TRAF6. We next analyzed the protein structures of RANK and constructed a 3D protein model of the extracellular region of klotho (Figure 5D). We used the ZDOCK program to explore the top 20 hot spots of RANK and klotho as an interface from 2000 complex structures (Figure 5E, Table 1). Finally, we identified the docking structure of the RANK/klotho complex (Figure 5F). To further identify the function of the RANK/klotho complex, we carried out co-immunoprecipitation (Co-IP) to detect the recruitment of TRAF6 by RANK activation in RAW264.7 cells with klotho overexpression/knockdown. We found that overexpression of klotho promoted the process in which TRAF6 was recruited and bound with activated RANK (Figure 5G), whereas knockdown of klotho significantly slowed this process (Figure 5H). Therefore, these data demonstrate that klotho binds to RANK, thus facilitating the interaction between RANK and TRAF6.

Table 1
| α-klotho | RANK | Cα distance (Å) | |||
|---|---|---|---|---|---|
| Residue | Site | Residue | Site | ||
| ASP | 545 | CYS | 152 | 7.76 | |
| ASP | 545 | THR | 153 | 7.51 | |
| PRO | 546 | THR | 153 | 6.92 | |
| VAL | 548 | THR | 153 | 7.51 | |
| TYR | 549 | GLN | 141 | 7.8 | |
| TYR | 549 | THR | 153 | 5.19 | |
| TYR | 549 | PRO | 154 | 6.12 | |
| TYR | 549 | CYS | 155 | 6.59 | |
| TYR | 549 | LEU | 156 | 6.49 | |
| LEU | 550 | GLY | 139 | 6.82 | |
| LEU | 550 | ALA | 140 | 7.23 | |
| LEU | 550 | GLN | 141 | 5.21 | |
| LEU | 550 | HIS | 142 | 6.88 | |
| LEU | 550 | THR | 153 | 5.92 | |
| LEU | 550 | PRO | 154 | 7.57 | |
| LEU | 550 | CYS | 155 | 7.02 | |
| LEU | 550 | LEU | 156 | 7.72 | |
| LEU | 550 | LYS | 169 | 7.07 | |
| LEU | 550 | CYS | 170 | 7.31 | |
| TRP | 551 | CYS | 155 | 6.64 | |
| TRP | 551 | LEU | 156 | 5.75 | |
| TRP | 551 | PHE | 159 | 7.48 | |
| TRP | 551 | CYS | 170 | 7.08 | |
| ASP | 552 | LEU | 156 | 7.17 | |
| LYS | 571 | PRO | 154 | 7.04 | |
| PRO | 572 | GLY | 137 | 6.83 | |
| PRO | 572 | PHE | 138 | 7.69 | |
| PRO | 572 | PRO | 154 | 6.86 | |
| CYS | 574 | ALA | 135 | 7.24 | |
| VAL | 575 | CYS | 134 | 6 | |
| VAL | 575 | ALA | 135 | 4.07 | |
| VAL | 575 | PRO | 136 | 6.57 | |
| VAL | 575 | GLY | 137 | 7.31 | |
| VAL | 575 | PHE | 138 | 6.62 | |
| ASP | 576 | CYS | 134 | 7.4 | |
| ASP | 576 | ALA | 135 | 7.22 | |
| ASP | 576 | CYS | 152 | 7.84 | |
| SER | 578 | THR | 132 | 6.1 | |
| SER | 578 | GLU | 133 | 6.82 | |
| SER | 578 | CYS | 134 | 7.86 | |
| ARG | 581 | THR | 132 | 7.63 | |
| ARG | 581 | GLU | 133 | 5.84 | |
| ARG | 581 | CYS | 134 | 7.15 | |
| ARG | 581 | ALA | 135 | 6.36 | |
| ARG | 581 | PHE | 164 | 6.4 | |
| PRO | 582 | GLU | 133 | 7.42 | |
| ILE | 584 | PHE | 164 | 7.87 | |
| THR | 585 | PHE | 164 | 6.88 | |
| ALA | 934 | CYS | 127 | 6.8 | |
| ALA | 934 | CYS | 128 | 7.05 | |
| ALA | 934 | ARG | 129 | 6.59 | |
| ASN | 935 | CYS | 128 | 7.72 | |
| ASN | 935 | ARG | 129 | 5.24 | |
| ASN | 935 | ARG | 130 | 6.28 | |
| GLN | 936 | ARG | 129 | 6.99 | |
| GLN | 936 | ARG | 130 | 6.04 | |
RANK, nuclear factor kappa B.
Discussion
The KL gene was initially identified as an anti-aging gene. Klotho mutant mice, in which the expression of klotho is disrupted, inherit a syndrome resembling human aging (28-30). In this study, we focused on the functional role and related mechanism of klotho in osteoclastogenesis. We found that klotho combined with RANK to promote the interactions between RANK and TRAF6, and subsequently activated the RANKL-induced NF-κB signaling pathway, ultimately facilitating osteoclastogenesis.
Klotho is encoded by the KL gene in humans (3). The extracellular domain is composed of 2 internal repeats and has homology to family 1 glycosidases. These 2 domains form a butterfly-shaped molecule on the surface of the cellular membrane (22); however, the intracellular domain has functional domains but the specific function is still unknown. Klotho mRNA is predominantly expressed in the kidneys, brain, and reproductive organs (3). One hallmark of aging-related phenotypes of klotho mutant mice is osteoporosis (3). When bone resorption exceeds bone formation, osteoporosis develops. The pathophysiology of osteopenia observed in kl/kl mice is characterized as low-turnover osteoporosis in which both bone formation and bone resorption are impaired, however, the decrease of bone formation is more significant (6). In the current study, we identified that the numbers of osteoblasts or osteoclasts per bone perimeter were decreased in femurs of klotho mutant mice, which is consistent with previous findings (Figure S1A,S1B). Moreover, we used RANKL to induce osteoclastogenesis in BMMs isolated from kl/kl mice and found that the lack of klotho showed impairment of osteoclastogenesis (Figure 1A-1D).
Studies have identified the presence of the klotho protein in osteocytes (31), and the disruption of klotho in osteocytes contributes to the osteoporotic bone phenotype in kl/kl mice, especially bone formation. However, the deficiency of klotho in osteocytes showed no significant effect on osteoclast resorption (32). Although bone is not the main distribution organ of klotho, recent studies provide new insights into the function of klotho expressed in bone cells, indicating that klotho may be expressed in different bone cells and functions as a regulator for original cells, such as osteocytes, osteoblastic cells and osteoclastic cells. Therefore, we measured the expression of klotho mRNA and protein during osteoclast differentiation. Notably, the results showed that klotho was indeed expressed in RANKL-induced BMMs or RAW264.7 cells and shared the same expression pattern changes with NFATc1 and c-Fos during osteoclastogenesis, which was elevated at the early phase of differentiation and decreased gradually. NFATc1 is an indispensable factor for osteoclastogenesis (17,18,28). Furthermore, c-Fos is a key transcription factor at the early stage during osteoclast differentiation that belongs to the activator protein-1 (AP-1) family (25). Studies have also identified that c-Fos is recruited to the NFATc1 promoter and is an indispensable factor for the early induction of NFATc1 for osteoclast differentiation (26,27). Our data suggested that klotho might be a regulator of osteoclastogenesis, thus we transfected the RAW264.7 cells with lentivirus vectors for klotho overexpression or klotho knockdown (Figure 2D-2G,2J). RAW 264.7 cells are generally used for studies of osteoclasts as the macrophage/pre-OC population, which will differentiate into functional osteoclasts upon stimulation of RANKL (33,34). We found that overexpression of klotho in RANKL-treated RAW264.7 cells promoted Osteoclastogenesis (Figures 2H,2I,3A,3B) and upregulated the expression of NFATc1 and c-Fos, while knockdown of klotho achieved the opposite results (Figures 2K,2L,3C,3D). These data identified that klotho is expressed in osteoclastic cells and upregulates NFATc1 and c-Fos to promote osteoclastogenesis.
As NFATc1 is a master transcriptional factor for osteoclast differentiation, our subsequent studies focused on the relationship between klotho and NFATc1 during osteoclastogenesis. In osteoclastic cells, several upstream signaling pathways of NFATc1 are activated by RANKL, including the NF-κB, MAPK, and AKT signalling pathways (35-37). We performed KEGG signaling pathway enrichment analysis using RANKL-induced BMMs isolated from kl/kl mice and WT mice and found that the NF-κB pathway was the most significantly activated pathway (Figure 4A-4C). Further tests for the phosphorylation of IκB and P65 in different RANKL-induced groups of cells identified that klotho functioned via activation of the NF-κB pathway (Figure 4D-4F), the effect of klotho on osteoclastogenesis in RANKL-treated RAW264.7 cells could be suppressed by the NF-κB pathway inhibitor BAY 11-7082 (Figure 4G,4H). Klotho mRNAs and proteins are localized at different sites in various cells. For example, in the kidney, klotho mRNAs and proteins are localized in the distal tubular cells (38). Moreover, it co-localizes with other proteins involved in tubular calcium reabsorption (39), suggesting that klotho proteins carry out their functions in various ways, especially through co-localizing with other proteins. To explore whether klotho affects the activation of the NF-κB pathway through regulating RANK and/or TRAF6, we firstly identified that klotho has no effect on the expression of RANK and TRAF6 (Figure 5A-5C). Further studies using the energy-based protein docking assay, which can be efficiently applied to identify interfaces and hot-spot residues in protein-protein complexes (Figure 5D-5F), revealed that klotho co-localized with RANK, suggesting it may regulate the interaction between RANK and TRAF6. Co-IP identified that TRAF6 was recruited and bound to RANK with RANKL stimulation, and klotho significantly facilitates this process (Figure 5G,5H), indicating that RANKL-induced recruitment of RANK with TRAF6 was positively regulated by klotho.
There are several limitations of the current study. Lentivirus transfection was not carried out in BMMs. In our pre-study, we found that cytotoxicity of lentivirus vectors led to the unstable proliferation of BMMs. Therefore, adenovirus may be an alternative vector for future study. One of the roles of klotho is to act as a co-receptor with fibroblast growth factor (FGF) receptor 1 (FGFR1) for FGF23. A previous study has found that external FGF23 also plays a role in osteoclastic cells. We performed some experiments and found that the expression of FGF23 was also present in osteoclastic cells during RANKL-stimulated osteoclastogenesis, although our subsequent experiments found that the expression of klotho and FGF23 was independent of each other (Figure S2). The specific role of FGF23 and whether it interacts with klotho during osteoclastogenesis was not fully elucidated in our research, and further study is needed.
Collectively, in this study, we demonstrated that klotho facilitates RANKL-induced osteoclastogenesis. Klotho co-localized with RANK, through which the interaction between RANK and TRAF6 was upregulated in RANKL-treated osteoclastic cells. The downstream NF-kB signaling pathway was thereby activated and ultimately increased the expression of NFATc1 and c-Fos. Therefore, klotho may be a promising therapeutic candidate for the treatment of osteoclast-associated osteopenic diseases.
Acknowledgments
The authors thank Qijie Dai of the Department of Orthopedics, Southwest Hospital, Third Military Medical University, for providing suggestions for experiments and helping with μCT scanning.
Funding: This work was supported by the National Natural Science Foundation of China (81902218 and 81672193).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-4332
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-4332
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-4332). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (No.: AMUWEC2021881) granted by the ethics board of The Third Military Medical University, in compliance with The Third Military Medical University institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature 2003;423:337-42. [Crossref] [PubMed]
- Takayanagi H, Kim S, Matsuo K, et al. RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-beta. Nature 2002;416:744-9. [Crossref] [PubMed]
- Kuro-o M, Matsumura Y, Aizawa H, et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 1997;390:45-51. [Crossref] [PubMed]
- Ohyama Y, Kurabayashi M, Masuda H, et al. Molecular cloning of rat klotho cDNA: markedly decreased expression of klotho by acute inflammatory stress. Biochem Biophys Res Commun 1998;251:920-5. [Crossref] [PubMed]
- Matsumura Y, Aizawa H, Shiraki-Iida T, et al. Identification of the human klotho gene and its two transcripts encoding membrane and secreted klotho protein. Biochem Biophys Res Commun 1998;242:626-30. [Crossref] [PubMed]
- Kawaguchi H, Manabe N, Chikuda H, et al. Cellular and molecular mechanism of low-turnover osteopenia in the klotho-deficient mouse. Cell Mol Life Sci 2000;57:731-7. [Crossref] [PubMed]
- Park JH, Lee NK, Lee SY. Current Understanding of RANK Signaling in Osteoclast Differentiation and Maturation. Mol Cells 2017;40:706-13. [PubMed]
- Fumoto T, Takeshita S, Ito M, et al. Physiological functions of osteoblast lineage and T cell-derived RANKL in bone homeostasis. J Bone Miner Res 2014;29:830-42. [Crossref] [PubMed]
- Xing L, Schwarz EM, Boyce BF. Osteoclast precursors, RANKL/RANK, and immunology. Immunol Rev 2005;208:19-29. [Crossref] [PubMed]
- Liu C, Walter TS, Huang P, et al. Structural and functional insights of RANKL-RANK interaction and signaling. J Immunol 2010;184:6910-9. [Crossref] [PubMed]
- Hsu H, Lacey DL, Dunstan CR, et al. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci U S A 1999;96:3540-5. [Crossref] [PubMed]
- Chung JY, Park YC, Ye H, et al. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J Cell Sci 2002;115:679-88. [Crossref] [PubMed]
- Feng X. Regulatory roles and molecular signaling of TNF family members in osteoclasts. Gene 2005;350:1-13. [Crossref] [PubMed]
- Liu W, Xu D, Yang H, et al. Functional identification of three receptor activator of NF-kappa B cytoplasmic motifs mediating osteoclast differentiation and function. J Biol Chem 2004;279:54759-69. [Crossref] [PubMed]
- Walsh MC, Kim GK, Maurizio PL, et al. TRAF6 autoubiquitination-independent activation of the NFkappaB and MAPK pathways in response to IL-1 and RANKL. PLoS One 2008;3:e4064 [Crossref] [PubMed]
- Wong BR, Josien R, Lee SY, et al. The TRAF family of signal transducers mediates NF-kappaB activation by the TRANCE receptor. J Biol Chem 1998;273:28355-9. [Crossref] [PubMed]
- Takayanagi H, Kim S, Koga T, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell 2002;3:889-901. [Crossref] [PubMed]
- Takanami-Ohnishi Y, Amano S, Kimura S, et al. Essential role of p38 mitogen-activated protein kinase in contact hypersensitivity. J Biol Chem 2002;277:37896-903. [Crossref] [PubMed]
- Kim K, Kim JH, Lee J, et al. Nuclear factor of activated T cells c1 induces osteoclast-associated receptor gene expression during tumor necrosis factor-related activation-induced cytokine-mediated osteoclastogenesis. J Biol Chem 2005;280:35209-16. [Crossref] [PubMed]
- Leaver-Fay A, Tyka M, Lewis SM, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol 2011;487:545-74. [Crossref] [PubMed]
- Alford RF, Leaver-Fay A, Jeliazkov JR, et al. The Rosetta All-Atom Energy Function for Macromolecular Modeling and Design. J Chem Theory Comput 2017;13:3031-48. [Crossref] [PubMed]
- Leman JK, Weitzner BD, Lewis SM, et al. Macromolecular modeling and design in Rosetta: recent methods and frameworks. Nat Methods 2020;17:665-80. [Crossref] [PubMed]
- Pierce BG, Wiehe K, Hwang H, et al. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 2014;30:1771-3. [Crossref] [PubMed]
- Padhorny D, Kazennov A, Zerbe BS, et al. Protein-protein docking by fast generalized Fourier transforms on 5D rotational manifolds. Proc Natl Acad Sci U S A 2016;113:E4286-93. [Crossref] [PubMed]
- Wang ZQ, Ovitt C, Grigoriadis AE, et al. Bone and haematopoietic defects in mice lacking c-fos. Nature 1992;360:741-5. [Crossref] [PubMed]
- Asagiri M, Sato K, Usami T, et al. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J Exp Med 2005;202:1261-9. [Crossref] [PubMed]
- Anderson DM, Maraskovsky E, Billingsley WL, et al. A homologue of the TNF receptor and its ligand enhance T-cell growth and dendritic-cell function. Nature 1997;390:175-9. [Crossref] [PubMed]
- Anamizu Y, Kawaguchi H, Seichi A, et al. Klotho insufficiency causes decrease of ribosomal RNA gene transcription activity, cytoplasmic RNA and rough ER in the spinal anterior horn cells. Acta Neuropathol 2005;109:457-66. [Crossref] [PubMed]
- Kawaguchi H, Manabe N, Miyaura C, et al. Independent impairment of osteoblast and osteoclast differentiation in klotho mouse exhibiting low-turnover osteopenia. J Clin Invest 1999;104:229-37. [Crossref] [PubMed]
- Mian IS. Sequence, structural, functional, and phylogenetic analyses of three glycosidase families. Blood Cells Mol Dis 1998;24:83-100. [Crossref] [PubMed]
- Rhee Y, Bivi N, Farrow E, et al. Parathyroid hormone receptor signaling in osteocytes increases the expression of fibroblast growth factor-23 in vitro and in vivo. Bone 2011;49:636-43. [Crossref] [PubMed]
- Komaba H, Kaludjerovic J, Hu DZ, et al. Klotho expression in osteocytes regulates bone metabolism and controls bone formation. Kidney Int 2017;92:599-611. [Crossref] [PubMed]
- Song H, Li X, Zhao Z, et al. Reversal of Osteoporotic Activity by Endothelial Cell-Secreted Bone Targeting and Biocompatible Exosomes. Nano Lett 2019;19:3040-8. [Crossref] [PubMed]
- Wu M, Chen W, Lu Y, et al. Gα13 negatively controls osteoclastogenesis through inhibition of the Akt-GSK3β-NFATc1 signalling pathway. Nat Commun 2017;8:13700. [Crossref] [PubMed]
- Tanaka S, Amling M, Neff L, et al. c-Cbl is downstream of c-Src in a signalling pathway necessary for bone resorption. Nature 1996;383:528-31. [Crossref] [PubMed]
- Mizukami J, Takaesu G, Akatsuka H, et al. Receptor activator of NF-kappaB ligand (RANKL) activates TAK1 mitogen-activated protein kinase kinase kinase through a signaling complex containing RANK, TAB2, and TRAF6. Mol Cell Biol 2002;22:992-1000. [Crossref] [PubMed]
- Wong BR, Besser D, Kim N, et al. TRANCE, a TNF family member, activates Akt/PKB through a signaling complex involving TRAF6 and c-Src. Mol Cell 1999;4:1041-9. [Crossref] [PubMed]
- Li SA, Watanabe M, Yamada H, et al. Immunohistochemical localization of Klotho protein in brain, kidney, and reproductive organs of mice. Cell Struct Funct 2004;29:91-9. [Crossref] [PubMed]
- Chang Q, Hoefs S, van der Kemp AW, et al. The beta-glucuronidase klotho hydrolyzes and activates the TRPV5 channel. Science 2005;310:490-3. [Crossref] [PubMed]
(English Language Editor: C. Betlzar)


