A novel prognostic signature for idiopathic pulmonary fibrosis based on five-immune-related genes
Introduction
Idiopathic pulmonary fibrosis (IPF) is a deadly interstitial lung disorder of unknown etiology (1). It is characterized by irreversible fibrogenesis in the lung parenchyma, leading to progressive respiratory function failure and eventually death (2,3). IPF is the most common interstitial lung disease and has the worst prognosis in pulmonary fibrosis (4). Nearly half of IPF patients die within 2–3 years after diagnosis (3,4), and the 5-year survival rate is less than 30% (5). IPF is a highly heterogeneous disease with a greatly variable natural history (6,7). The course of this disease in an individual patient is difficult to predict (4,8); some patients with IPF experience rapid decline, while others experience much slower development (3,8). For a long time, the lack of effective prognostic indicators has made it difficult to accurately track and evaluate the prognosis of IPF, which has led to the poor prognosis of IPF to a certain extent. Hence, the development of applicable prognostic signatures is urgently needed for the clinical treatment of IPF.
The pathophysiological pathogenesis of IPF involves aberrant transcription and gene expression (9-14). Molecular genomic features based on lung tissue have been used to predict the development of IPF (15,16). Though previous studies have identified some genes and pathways may play an important role in the occurrence and development of IPF, and may be expected to be biomarkers or therapeutic targets for the diagnosis of IPF (17,18). However, the lack of verification of survival information is the biggest short board in these papers. Meanwhile, the resources required to perform a lung biopsy and the risks associated with the procedure limit the applicability of such genomic features. Molecular models have also been established based on peripheral blood mononuclear cell (PBMC) transcription profile data to predict the disease state of IPF (19,20). However, in the absence of lung biopsies, it is difficult to explain the correlation between abnormal PBMC transcription and pulmonary fibrosis course. Bronchoalveolar lavage (BAL) is a method of obtaining alveolar surface lining fluid with fiberoptic bronchoscopy for evaluating inflammation, immune cells, and soluble substances. BAL plays a vital role in assisting IPF diagnosis and has been recommended as the auxiliary diagnostic reference by the American Thoracic Society (ATS) (21). The advantages of utilizing the gene expression profiles of BAL cells to depict the molecular features of IPF include lung localization, ease of accessibility, and dynamic assessment of disease status through longitudinal sample collection. Previous studies have revealed that Innate and adaptive immune responses disorders possess an important role in the pathogenesis of lung fibrosis (22). The differentially-expressed immune-related genes (IRGs) also have been reported associated with the development of IPF (23,24). The immPort database is funded by the National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases (NIAID), Health and Human Services (HHS) in support of the NIH mission to share data with the public. It provides information about the immune-related genes of humans. Therefore, using the GSE70866 gene expression data set of the Gene Expression Synthesis (GEO) database and the IRGs list of the ImmPort database, we aim to combine the survival information of IPF patients to establish a new molecular genome feature screening from IRGs, to predict the prognosis of IPF patient. We present the following article in accordance with the STARD reporting checklist (available at https://dx.doi.org/10.21037/atm-21-4545).
Methods
Acquisition and analysis of datasets
Microarray profile data from the GSE70866 gene expression dataset were downloaded from the GEO (http://www.ncbi.nlm.nih.gov/geo/) database. The platform was a GPL14550 Agilent-028004 SurePrint G3 Human GE 8x60K Microarray (Agilent Technologies Inc., California, U.S.). A total of 132 BALF samples, including 20 samples from healthy individuals and 112 samples from IPF patients, were used to analyze the microarray data. All 112 IPF patients had detailed sociodemographic characteristics and complete survival information. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
The criteria of differentially-expressed genes (DEGs) and differentially-expressed immune-related genes (IRGs)
The filtration of DEGs was performed in 112 IPF patients versus healthy individuals. In this study, DEGs between IPF and healthy individuals were defined using a log2 fold change (FC) >1 and an adjusted P value (adj. P) <0.05 as thresholds. A total of 1,793 IRGs were obtained from the ImmPort (https://www.immport.org/shared/genelists) database. Taking the intersection through the Venn algorithm (http://bioinformatics.psb.ugent.be/webtools/Venn/), 52 differentially-expressed IRGs were filtered, which remained and were used as candidates for subsequent analysis.
Construction and validation of the prognostic IRG-based signature
The 112 included patients were randomly divided into a training cohort (50%) and validation cohort (50%) using the random numbers method. The construction of prognostic gene-based signatures was carried out in the training cohort, and verification was performed in the verification cohort. Univariate Cox regression analysis was used to screen for immune genes that were significantly associated with prognosis, with a cut-off of P<0.05. Next, multivariate Cox-regression analysis was performed on the training cohort to further determine the best prognostic IRG signature using the “survival” package (URL: https://github.com/therneau/survival) in R software (version 4.0.3) (URL: https://cran.r-project.org/mirrors.html), with a cut-off of P<0.05. The formula of IPF patient’s risk score was established as follows: score = sum (each gene’s expression × corresponding coefficient). The patients were stratified into high-risk and low-risk groups based on the median value of the risk score. Based on the risk score groups, survival differences between high-risk and low-risk groups were carried out with the “survival” R package (URL: https://github.com/therneau/survival).
Statistical analysis
Baseline characteristics such as age, sex, race, days to death, and vital status were collected. Continuous variables were reported as the mean (± standard deviation) and compared using the Student’s t-test. Categorical variables were reported as counts n (%) and compared using the chi-square test. The comparison of sociodemographic features between the training and validation cohorts was carried out using GraphPad Prism (version 7.0; GraphPad Software, La Jolla, CA, USA).
The other statistical analyses were carried out using R software (version 4.0.3) (URL: https://cran.r-project.org/mirrors.html) and considered significant when the corresponding P<0.05. The adjusted P<0.05 was used for screening DEGs, and P<0.05 was used as a significance threshold in the remaining statistical analyses. The analysis of DEGs was conducted by utilizing the “limma” package (URL: http://www.strimmerlab.org/software/st/). Univariate Cox regression analysis was used to screen for DEGs that were significantly associated with overall survival (OS). Multivariate Cox regression analysis was performed on the training cohort to further determine the best prognostic IRG model. A multivariate Cox regression model was conducted for the variables with P<0.05 in the univariate analyses. A gene-based signature was built with the coefficients of each factor in the multivariate Cox analysis. The “survival” package (URL: https://github.com/therneau/survival) calculated the survival curve function, and the “survminer” package (URL: https://mirror.lzu.edu.cn/CRAN/bin/windows/contrib/4.0/survminer_0.4.9.zip) executed the visualization. The heat map was drawn using the “pheatmap” (pretty heatmap) package (URL: https://mirror.lzu.edu.cn/CRAN/bin/windows/contrib/4.0/pheatmap_1.0.12.zip). The volcano map was drawn using the “ggplot2” package (URL: https://cran.r-project.org/web/packages/ggplot2movies/index.html).
Results
Baseline characteristics of patient with IPF
Table 1 summarizes the sociodemographic information of the included IPF patients. A total of 112 IPF patients were identified, with a median age of 69.5 (±10.1) years. IPF was more common in older populations (67.0% of patients were older than 65 years versus 33.0% of patients less than 6 years). The incidence of IPF was higher in men than in women (83.0% male patients versus 17.0% female patients).
Table 1
| Characteristics | Total (n=112) | Training cohort (n=56) | Validation cohort (n=56) | P value |
|---|---|---|---|---|
| Age, mean (± SD) | 67.97 (±10.1) | 67.0 (±10.4) | 69.0 (±9.7) | 0.300 |
| Age, n (%) | ||||
| <65 | 37 (33.0) | 18 (32.1) | 19 (34.0) | |
| ≥65 | 75 (67.0) | 38 (67.9) | 37 (66.0) | 0.841 |
| Gender, n (%) | ||||
| Female | 19 (17.0) | 7 (12.5) | 12 (21.4) | |
| Male | 93 (83.0) | 49 (87.5) | 44 (78.6) | 0.208 |
| Days to death, mean (± SD) | 698.1 (±555.9) | 656.7 (±551.9) | 739.5 (±561.7) | 0.433 |
| Vital status, n (%) | ||||
| Alive | 36 (32.1) | 20 (35.7) | 16 (28.6) | |
| Dead | 76 (67.9) | 36 (64.3) | 40 (71.4) | 0.418 |
| Sample contact country, n (%) | ||||
| Germany | 112 (100.0) | 56 (100.0) | 56 (100.0) | NA |
SD, standard deviation.
These 112 IPF patients were randomly divided into training (50%) and validation (50%) cohorts, with 56 patients in each group. No significant differences between the two cohorts were observed in terms of age, sex, days to death, and vital status (P>0.05). Qualified survival information for all of the included IPF patients was available for further analysis.
Identification of DEGs
DEGs of the IPF and healthy individuals from the GPL14550 platform of the GSE70866 gene expression dataset were analysed using the “limma” package. In this dataset, a total of 379 DEGs met the criteria, of which 207 genes were upregulated and 172 genes were downregulated (Table S1). Figure 1A is a volcano map of 379 DEGs in the IPF group compared to the healthy individuals group. The profiling of all the DEGs is shown in Figure 1B and presented in the form of a non-cluster analysis expression heatmap. SPP1, PPBP, and MMP7 were the top three most significantly upregulated genes in the IPF group, while NALCN, C8B, and ITIH5 were the three most downregulated genes in the IPF group.

Identification of differential expression IRGs
Combining the results of DEGs (Table S1) and the IRGs from the ImmPort database, 52 differentially expressed IRGs were identified. A volcano map was constructed to present the differential expression of all IRGs (Figure 2A). Figure 2B shows the expression of the 52 differential IRGs in the form of a heatmap. SPP1, PPBP, TUBB3, CCL2, and S100A12 were the five most significantly upregulated IRGs, while the top five downregulated IRGs were PTGER3, CD40LG, CAMP, IGF1, and CXCL9.

Prognostically relevant IRGs filtration
Prognostically relevant IRGs for IPF were selected based on the results of univariate Cox regression analysis. A forest plot was drawn to show the 37 obtained prognostically relevant IRGs, including prognostically protective IRGs such as RORA [hazard ratio (HR): 0.613, 95% confidence interval (CI): (0.474–0.794)] and ICOS [HR: 0.672, 95% CI: (0.560–0.809)] (Figure 3). Conversely, MPO [HR: 1.287, 95% CI: (1.139–1.454)], RNASE3 [HR: 1.711, 95% CI: (1.338-2.188)], PDGFA [HR: 1.228, 95% CI: (1.030–1.465)], PPBP [HR: 1.154, 95% CI: (1.002–1.330)], and FABP3 [HR: 1.522, 95% CI: (1.216–1.905)] were prognostic factors of worse survival (Figure 3).

An IRGs prognostic model of IPF
Multivariate Cox regression analysis was performed based on 37 prognostic factors of OS to establish a model to predict the outcomes of IPF patients. CXCL14, SLC40A1, RNASE3, CCR3, and RORA were ultimately identified to build a five-IRG-based prognostic signature to predict the survival time of patients with IPF in the training cohort.
Figure 4A-4E shows the survival outcomes of IPF patients stratified by CXCL14, SLC40A1, RNASE3, CCR3, and RORA. The survival curve revealed that IPF patients with higher expression levels of CXCL14, SLC40A1, RNASE3, and CCR3 had much worse survival outcomes. Patients with a relatively lower expression of RORA had markedly longer OS.

Detailed results of the multivariate Cox regression analysis, including coefficients, P values, hazard ratios, etc., are provided in Table S2. Accordingly, the patient’s risk score representing the risk for OS was calculated as follows: risk score = 0.1970 × expression value of CXCL14 + 0.3280 × expression value of SLC40A1 + 0.5852× expression value of RNASE3 + 0.2802 × expression value of CCR3 − 0.6504 × expression value of RORA. According to the median risk score, IPF patients were divided into high- and low-risk groups. Individuals with risk scores beyond 0.711 were recognized as high-risk; otherwise, they were considered low-risk (Figure 5A, Table S3). There was a significant decrease in the OS of IPF patients as the risk score increased (Figure 5B). Figure 5C displays the expression level of the five IRGs between the high- and low-risk groups. As shown in Figure 5C, CXCL14, SLC40A1, RNASE3, and CCR3 were more highly expressed, while RORA expression exhibited relatively lower expression in the high-risk IPF patients than in the low-risk individuals. The survival curve constructed by the five-IRG-based prognostic signature in the training cohort showed that there was an extremely significant difference between the high- and low-risk groups (Figure 6A). A validation cohort was utilized to verify the five-IRG-based signature, and notable differential survival outcomes were observed between the high- and low-risk groups (Figure 6B). The area under curves (AUC) of the five-IRG-based prognostic signature for IPF in the training model was 0.858 (Figure 6C). The AUC of this predictive five-gene-based signature in the validation was 0.837 (Figure 6D), indicating that this predictive signature could be trusted.


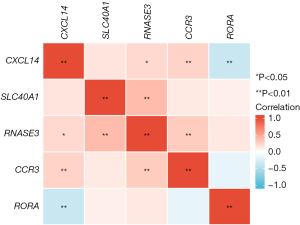
Correlation expression map of the five genes included in the predictive signature
A correlation map of the five included prognostic IRGs expression levels is described in Figure 7. The strongest expression correlations were observed between RNASE3 and SLC40A1 (P<0.01, r=0.394), as well as between RORA and CXCL14 (P<0.01, r=−0.355). Meanwhile, the expression level of CCR3 was significantly positively correlated with the expression of CXCL14 (P<0.01, r=0.258). There was an intimate positive association between RNASE3 and CCR3 (P<0.01, r=0.293).
Discussion
IPF is the most prevalent subtype of interstitial lung disease (ILD) worldwide (25). However, it has the poorest prognosis among the various ILD subtypes, with a median survival of 2–3 years after diagnosis (3,4). Lung transplantation is the only intervention that has been shown to prolong survival for patients with IPF (26). Pirfenidone and nintedanib have emerged as effective therapies that can significantly slow the decline in forced vital capacity (FVC) and disease progression in IPF patients (27,28). However, the prognosis of IPF remains unfavourable. The poor prognosis of IPF is partly due to a lack of effective prognostic biomarkers to guide treatment. Without the ability to forecast disease progression, it is difficult to determine which IPF patients are likely to benefit from new therapies or lung transplantation. Therefore, we constructed a molecular genomic signature to predict the prognosis of IPF patients using the GSE70866 gene expression dataset from the GEO database.
Previous studies have revealed that the immune system possesses an actual effect on the IPF process (22,29,30). All stages of fibrogenesis are accompanied by innate and adaptive immune responses (22). More importantly, increasing evidence has appeared over the last few years establishing the meaningful role of IRGs in the pathogenesis and treatment of lung fibrosis (23,24,31,32). It has been shown that regulating the expression of IRGs can ameliorate pulmonary fibrogenesis in bleomycin-induced (BLM-induced) mouse models (31,32). Furthermore, data from clinical trials of newly developed drugs for the treatment of IPF have demonstrated the active role of IRG-targeting drugs in slowing disease progression. For instance, IRG-targeting drugs have been shown to play a positive role in reducing fibrogenesis (33). These previous studies highlight the importance of IRGs in the pathophysiological mechanism of IPF. In the present study, we were interested in the role of IRGs in the prognosis of IPF.
In total, 112 IPF patients and 20 healthy individuals were included in our study. The included IPF patients were predominantly older males (aged >65 years old). This demographic feature, as well as the fact that the prevalence of IPF is higher in men than in women, are consistent with previous studies (1,3). In this comparative microarray profile of an IPF cohort versus a healthy individual cohort, a total of 379 DEGs were identified. The genes involved in encoding extracellular matrix (ECM) components, tissue architecture remodeling, and ECM accumulation (SPP1, MMP7, MMP10, CCL2, and ITGB3) were observed to be significantly upregulated (34-37). Of the 379 DEGs, 52 were filtered as IRGs based on the ImmPort database. Next, 37 of these 52 differentially-expressed IRGs were recognized as significant prognostic biomarkers for patients with IPF. More than 70% of the differentially-expressed IRGs had notable associations with survival. Our results further suggested that there was a close association between IRGs and the progression of IPF, which was consistent with previous studies. Based on these findings, a five IRG-based prognostic signature (CXCL14, SLC40A1, RNASE3, CCR3, and RORA), was built in the training cohort in this study. This signature presented an excellent predictive prognostic effect, with an AUC value of 0.858. In addition, the risk score was significantly different between the high- and low-risk groups. Meanwhile, the risk score was significantly correlated with the OS of IPF patients. CXCL14, SLC40A1, CXCL14, and CCR3 were differentially-upregulated genes between IPF patients and healthy individuals. The expression levels of these four genes in the high-risk IPF group were significantly higher than those in the low-risk group. RORA was detected at a lower expression level in the healthy individuals group compared to the IPF group. Consistently, the expression level of RORA was lower in the high-risk IPF group than in the low-risk group.
Fibroblast foci represent the main pathogenic lesions of IPF, including abnormally activated fibroblasts and myofibroblasts. Myofibroblasts are the main effector cells of IPF. They can secrete a large amount of ECM protein and promote the abnormal hardening of ECM, which leads to the remodeling of lung structure and the gradual loss of lung function (38-40). Previous studies have confirmed that knockdown of CXCL14 could inhibit lung fibrogenesis by suppressing lung fibroblasts proliferation and downregulating MMP2/9 (31). Zagai et al. found eosinophil cationic protein (ECP, also known as RNASE3) could stimulate human lung fibroblasts to secrete extracellular matrix, thereby leads to airway fibrosis (41). The concentration of RNASE3 in bronchoalveolar lavage fluid (BALF) is markedly increased in IPF patients compared with healthy individuals and is highly correlated with acute exacerbation during the preceding 3- to 6-month period (42,43). CCR3 can increase the activation, migration and proliferation ability of lung fibroblasts, and the ability of myofibroblasts to secrete ECM protein (44,45). In addition, CCR3 is notably expressed in the lungs of BLM-induced mice and is expressed not only by eosinophils but also by neutrophils (44). CCR3 plays a key role in the recruitment of granulocytes and is an important suppressor of fibrogenesis in BLM-treated lungs (44). These studies on the pathophysiological mechanisms between IPF and CXCL14, RNASE3, and CCR3 increase the credibility of the signature constructed in our study. Our research also showed that there is a meaningful correlation between the expression of RNASE3 and CCR3. Meanwhile, a significant expression correlation between CXCL14 and CCR3 was also observed in this study. For the SLC40A1 and RORA, no relevant studies have been conducted to determine the association with lung fibrosis. We first reported that there may be some potential associations between the pathological mechanism of IPF and SLC40A1 along with RORA. The specific pathophysiological mechanism is worthy of further study.
Finally, we evaluated the performance of the genomic signature in the validation cohort. The signature showed an equally excellent ability to distinguish between high- and low-risk patient groups. The AUC value of the Receiver Operating Characteristic Curve (ROC) curve was 0.837, demonstrating the potential applicability of our findings for real-world use.
While the genomic model developed in this study was successfully validated, there were still some potential limitations that should be noted. Firstly, this research was based on the gene expression profiles from the GEO database. Due to the difficult of recruitment of a large number of IPF patients, no validation of the 5 genes in real world data in this paper. Also, the IPF patients included in this study were all from Germany. Thus, our results might only represent patients in Germany and might not applicable to all IPF patients worldwide. Finally, due to limited data on treatment, our study did not subgroup IPF patients according to the different treatment choices. Consequently, the reliability and accuracy of our results might be affected and needs to be re-evaluated by future studies.
Conclusions
In conclusion, our study identified a novel five-IRG-based signature that is a reproducible predictor of outcome in IPF patients. This novel signature benefits the personalized management of patients with IPF. Furthermore, this finding provides new insights into the relationship between the immune system and IPF, offering incremental clinical value for IPF prognosis and therapy.
Acknowledgments
We acknowledge GEO database for providing their platforms and contributors for uploading their meaningful datasets.
Funding: This study was supported by grants from the Henan Provincial Key Laboratory for Interstitial Lung Disease and Lung Transplantation.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-4545
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-4545). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The data used in this study was derived from a public database, and thus, no ethical approval was needed.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Raghu G, Remy-Jardin M, Myers JL, et al. Diagnosis of Idiopathic Pulmonary Fibrosis. An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline. Am J Respir Crit Care Med 2018;198:e44-68. [Crossref] [PubMed]
- Selman M, King TE, Pardo A, et al. Idiopathic pulmonary fibrosis: prevailing and evolving hypotheses about its pathogenesis and implications for therapy. Ann Intern Med 2001;134:136-51. [Crossref] [PubMed]
- Raghu G, Collard HR, Egan JJ, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med 2011;183:788-824. [Crossref] [PubMed]
- Mura M, Porretta MA, Bargagli E, et al. Predicting survival in newly diagnosed idiopathic pulmonary fibrosis: a 3-year prospective study. Eur Respir J 2012;40:101-9. [Crossref] [PubMed]
- Vancheri C, du Bois RM. A progression-free end-point for idiopathic pulmonary fibrosis trials: lessons from cancer. Eur Respir J 2013;41:262-9. [Crossref] [PubMed]
- Kim HJ, Perlman D, Tomic R. Natural history of idiopathic pulmonary fibrosis. Respir Med 2015;109:661-70. [Crossref] [PubMed]
- Kim DS, Collard HR, King TE Jr. Classification and natural history of the idiopathic interstitial pneumonias. Proc Am Thorac Soc 2006;3:285-92. [Crossref] [PubMed]
- Ley B, Collard HR, King TE Jr. Clinical course and prediction of survival in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2011;183:431-40. [Crossref] [PubMed]
- Selman M, López-Otín C, Pardo A. Age-driven developmental drift in the pathogenesis of idiopathic pulmonary fibrosis. Eur Respir J 2016;48:538-52. [Crossref] [PubMed]
- Bueno M, Lai YC, Romero Y, et al. PINK1 deficiency impairs mitochondrial homeostasis and promotes lung fibrosis. J Clin Invest 2015;125:521-38. [Crossref] [PubMed]
- Prasse A, Binder H, Schupp JC, et al. BAL Cell Gene Expression Is Indicative of Outcome and Airway Basal Cell Involvement in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med 2019;199:622-30. [Crossref] [PubMed]
- Caporarello N, Meridew JA, Jones DL, et al. PGC1α repression in IPF fibroblasts drives a pathologic metabolic, secretory and fibrogenic state. Thorax 2019;74:749-60. [Crossref] [PubMed]
- McDonough JE, Kaminski N, Thienpont B, et al. Gene correlation network analysis to identify regulatory factors in idiopathic pulmonary fibrosis. Thorax 2019;74:132-40. [Crossref] [PubMed]
- DePianto DJ, Chandriani S, Abbas AR, et al. Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis. Thorax 2015;70:48-56. [Crossref] [PubMed]
- Boon K, Bailey NW, Yang J, et al. Molecular phenotypes distinguish patients with relatively stable from progressive idiopathic pulmonary fibrosis (IPF). PLoS One 2009;4:e5134 [Crossref] [PubMed]
- Korfei M, von der Beck D, Henneke I, et al. Comparative proteome analysis of lung tissue from patients with idiopathic pulmonary fibrosis (IPF), non-specific interstitial pneumonia (NSIP) and organ donors. J Proteomics 2013;85:109-28. [Crossref] [PubMed]
- Xu Z, Mo L, Feng X, et al. Using bioinformatics approach identifies key genes and pathways in idiopathic pulmonary fibrosis. Medicine (Baltimore) 2020;99:e22099 [Crossref] [PubMed]
- Min F, Gao F, Liu Z. Screening and further analyzing differentially expressed genes in acute idiopathic pulmonary fibrosis with DNA microarray. Eur Rev Med Pharmacol Sci 2013;17:2784-90. [PubMed]
- Li FJ, Surolia R, Li H, et al. Autoimmunity to Vimentin Is Associated with Outcomes of Patients with Idiopathic Pulmonary Fibrosis. J Immunol 2017;199:1596-605. [Crossref] [PubMed]
- Huang Y, Ma SF, Vij R, et al. A functional genomic model for predicting prognosis in idiopathic pulmonary fibrosis. BMC Pulm Med 2015;15:147. [Crossref] [PubMed]
- Meyer KC, Raghu G, Baughman RP, et al. An official American Thoracic Society clinical practice guideline: the clinical utility of bronchoalveolar lavage cellular analysis in interstitial lung disease. Am J Respir Crit Care Med 2012;185:1004-14. [Crossref] [PubMed]
- Heukels P, Moor CC, von der Thüsen JH, et al. Inflammation and immunity in IPF pathogenesis and treatment. Respir Med 2019;147:79-91. [Crossref] [PubMed]
- Cecchini MJ, Hosein K, Howlett CJ, et al. Comprehensive gene expression profiling identifies distinct and overlapping transcriptional profiles in non-specific interstitial pneumonia and idiopathic pulmonary fibrosis. Respir Res 2018;19:153. [Crossref] [PubMed]
- Walsh SM, Worrell JC, Fabre A, et al. Novel differences in gene expression and functional capabilities of myofibroblast populations in idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2018;315:L697-710. [Crossref] [PubMed]
- Lederer DJ, Martinez FJ. Idiopathic Pulmonary Fibrosis. N Engl J Med 2018;378:1811-23. [Crossref] [PubMed]
- George PM, Patterson CM, Reed AK, et al. Lung transplantation for idiopathic pulmonary fibrosis. Lancet Respir Med 2019;7:271-82. [Crossref] [PubMed]
- Richeldi L, Costabel U, Selman M, et al. Efficacy of a tyrosine kinase inhibitor in idiopathic pulmonary fibrosis. N Engl J Med 2011;365:1079-87. [Crossref] [PubMed]
- Taniguchi H, Ebina M, Kondoh Y, et al. Pirfenidone in idiopathic pulmonary fibrosis. Eur Respir J 2010;35:821-9. [Crossref] [PubMed]
- Drakopanagiotakis F, Wujak L, Wygrecka M, et al. Biomarkers in idiopathic pulmonary fibrosis. Matrix Biol 2018;68-69:404-21. [Crossref] [PubMed]
- O'Dwyer DN, Ashley SL, Moore BB. Influences of innate immunity, autophagy, and fibroblast activation in the pathogenesis of lung fibrosis. Am J Physiol Lung Cell Mol Physiol 2016;311:L590-601. [Crossref] [PubMed]
- Li L, Li Q, Wei L, et al. Chemokine (C-X-C motif) ligand 14 contributes to lipopolysaccharide-induced fibrogenesis in mouse L929 fibroblasts via modulating PPM1A. J Cell Biochem 2019;120:13372-81. [Crossref] [PubMed]
- Mehrad B, Burdick MD, Strieter RM. Fibrocyte CXCR4 regulation as a therapeutic target in pulmonary fibrosis. Int J Biochem Cell Biol 2009;41:1708-18. [Crossref] [PubMed]
- Lukey PT, Harrison SA, Yang S, et al. A randomised, placebo-controlled study of omipalisib (PI3K/mTOR) in idiopathic pulmonary fibrosis. Eur Respir J 2019;53:1801992 [Crossref] [PubMed]
- Insua-Rodríguez J, Pein M, Hongu T, et al. Stress signaling in breast cancer cells induces matrix components that promote chemoresistant metastasis. EMBO Mol Med 2018;10:e9003 [Crossref] [PubMed]
- Rohani MG, Parks WC. Matrix remodeling by MMPs during wound repair. Matrix Biol 2015;44-46:113-21. [Crossref] [PubMed]
- Kuo CS, Pavlidis S, Zhu J, et al. Contribution of airway eosinophils in airway wall remodeling in asthma: Role of MMP-10 and MET. Allergy 2019;74:1102-12. [Crossref] [PubMed]
- Liu A, Liu Y, Li B, et al. Role of miR-223-3p in pulmonary arterial hypertension via targeting ITGB3 in the ECM pathway. Cell Prolif 2019;52:e12550 [Crossref] [PubMed]
- Guillotin D, Taylor AR, Platé M, et al. Transcriptome analysis of IPF fibroblastic foci identifies key pathways involved in fibrogenesis. Thorax 2021;76:73-82. [Crossref] [PubMed]
- Richeldi L, Collard HR, Jones MG. Idiopathic pulmonary fibrosis. Lancet 2017;389:1941-52. [Crossref] [PubMed]
- Martinez FJ, Collard HR, Pardo A, et al. Idiopathic pulmonary fibrosis. Nat Rev Dis Primers 2017;3:17074. [Crossref] [PubMed]
- Zagai U, Dadfar E, Lundahl J, et al. Eosinophil cationic protein stimulates TGF-beta1 release by human lung fibroblasts in vitro. Inflammation 2007;30:153-60. [Crossref] [PubMed]
- Fujimoto K, Kubo K, Yamaguchi S, et al. Eosinophil activation in patients with pulmonary fibrosis. Chest 1995;108:48-54. [Crossref] [PubMed]
- Birring SS, Parker D, McKenna S, et al. Sputum eosinophilia in idiopathic pulmonary fibrosis. Inflamm Res 2005;54:51-6. [Crossref] [PubMed]
- Huaux F, Gharaee-Kermani M, Liu T, et al. Role of Eotaxin-1 (CCL11) and CC chemokine receptor 3 (CCR3) in bleomycin-induced lung injury and fibrosis. Am J Pathol 2005;167:1485-96. [Crossref] [PubMed]
- Puxeddu I, Bader R, Piliponsky AM, et al. The CC chemokine eotaxin/CCL11 has a selective profibrogenic effect on human lung fibroblasts. J Allergy Clin Immunol 2006;117:103-10. [Crossref] [PubMed]
(English Language Editor: B. Draper)