
A narrative review of research progress on FoxM1 in breast cancer carcinogenesis and therapeutics
Introduction
Breast cancer is widely seen as one of the most common malignant tumors in women, and is also the leading cause of cancer-related deaths among female tumor patients worldwide (1,2). In the past two decades, a large number of epidemiological surveys published in different regions of the world have shown that the incidence and mortality of breast cancer have increased notably (3). On account of the advancement and diversity of treatment methods, the mortality rate of breast cancer patients has reduced strikingly, but the recurrence and metastasis of tumors are still the central cause of death (4). In addition, the emergence of traditional drug resistance has also brought great difficulties for clinicians in the treatment of breast cancer patients, which directly affects the survival time of patients (5). Consequently, further understanding of the pathogenesis of breast cancer will help to improve the treatment and prognosis of breast cancer patients.
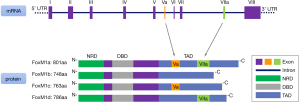
The human Fox gene family includes at least 40 subfamily members, such as FoxM1. Their common feature is that they have an evolutionary conserved homologous DNA binding region composed of 110 amino acids, which is also called a “wing-like helix” structure (6). FoxM1, also known as Trident, WIN, FKHL16, MPP2, and HFH-11, is a member of the forkhead box (Fox) transcription factor superfamily (7,8). The gene encoding human FoxM1 is located on chromosomal band 12p13.3, with a total length of 19.47kb and 10 exons (9). According to the different splices of exons Va and VIIa, FoxM1 is divided into 4 subtypes: FoxM1a, FoxM1b, FoxM1c, and FoxM1d. Each includes a N-terminal repressor region, a C-terminal transactivation region, and a highly conserved DNA binding region (1,10). FoxM1a retains both exons Va and VIIa sequences, whereas FoxM1b lacks both the exons sequences (11). FoxM1c only contains exon Va sequence, while FoxM1d only includes the exon VIIa sequence (Figure 1) (12). FoxM1a and FoxM1d do not directly play a role in transcriptional regulation and are predominantly distributed in the cytoplasm (13). In contrast, FoxM1b and FoxM1c are mainly located in the nucleus with transcriptional regulatory functions (14). Extensive studies have demonstrated that FoxM1b is expressed at elevated levels in most human tumor cells, and exhibits a greater transforming capability than FoxM1c (15,16).
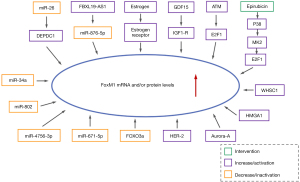
As a proliferation-specific transcription factor, FoxM1 is implicated in the regulation of various cellular activities, such as inflammation, apoptosis, drug resistance, metabolism, DNA damage repair, stem cell renewal, tissue regeneration, angiogenesis, metastasis, and maintenance of the integrity of mitotic spindles (17-19). The elevated expression of FoxM1 is closely related to cell division and proliferation, and also has the activities of binding DNA, other proteins, and other protein kinases (17,20). The high expression of FoxM1 has been detected in various types of human cancer cells (such as ovarian cancer, gastric cancer, pancreatic cancer, colorectal cancer), and the elevated expression of FoxM1 can promote the malignant characteristics of tumor cells, which indicates that FoxM1 is closely connected with tumorigenesis (21-24). FoxM1 is also an advanced cell cycle gene, which plays a vital role in tumor occurrence, invasion, and metastasis in breast cancer (20). Besides, many studies have revealed that FoxM1 is highly expressed in breast cancer (Figure 2), which plays a significant role in the prognosis and chemotherapy resistance of breast cancer patients (20). Thus, this review will concentrate on the recent studies investigating the role of this FoxM1 including initiation, proliferation, angiogenesis, invasion, metastasis and drug resistance of breast cancer, as well as its predictive value of FoxM1 as a prognostic biomarker and potential therapeutic target for breast cancer. Additionally, this review also briefly elaborates the link between this FoxM1 and non-coding RNAs in breast cancer, which provide predictive biomarkers and therapeutic intervention targets for breast cancer.
We present the following article in accordance with the Narrative Review reporting checklist (available at https://dx.doi.org/10.21037/atm-21-5271).
Roles of FoxM1 in breast cancer initiation
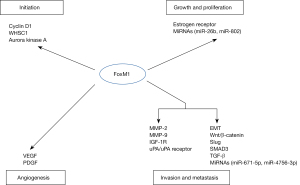
More and more published research articles have shown that FoxM1 plays a basic role in tumorigenesis, which is mostly correlated with the regulation of cell cycle progression (25,26). It is important that FoxM1 is a pivotal regulator in the cell cycle of the G1 phase to S phase and G2 phase to M phase via inducing the expression of cell cycle-related factors (27). FoxM1 expression varies with cell cycle stage, enhancing during S phase and peaking at G2-M under normal physiological conditions (28). Studies have shown that FoxM1 regulates the expression of a number of cell cycle proteins such as cyclin D1 (29). Cyclin D1 plays a key role in the G1 phase, G1/S transition, and oncogenesis. It was verified that FoxM1 was obviously up-regulated in triple-negative breast cancer (TNBC), while the knockdown of FoxM1 by RNA interference (siRNA) in TNBC cells contributed to a marked reduction in cyclin D1 expression, with its inhibition disrupting the initiation of breast cancer (30). Available statistics demonstrated that FoxM1 also facilitated the G2-M transition and the M phase through influencing regulators such as cyclin A2, cyclin B, cell division cycle 25B, polo-like kinase 1, Aurora kinase A, centromere protein (CENP)-A, CENP-B, and CENP-F (31-33). A study by Yang et al. (34) demonstrated that Aurora kinase A stabilized FoxM1 via attenuating its ubiquitin in late M phase and early G1 phase of the cell cycle, thereby promoting the proliferation and progression of TNBC cells. In addition, a research by Zhang et al. (35) elucidated that the expression of FoxM1 was regulated by Wolf-Hirschhorn syndrome candidate gene-1 (WHSC1), and the elevated WHSC1 expression induced the up-regulation of FoxM1 expression. Up-regulation of FoxM1 further led to more localization of β-catenin to the nucleus, resulting in overexpression of the downstream transcriptional activation of matrix metalloproteinase (MMP)-2 and MMP-9, which can give rise to the occurrence of breast cancer (Figure 3).
Roles of FoxM1 in breast cancer growth and proliferation
FoxM1, known as a proliferation-specific gene, is closely correlated with the growth and proliferation of various tumor cells (36,37). In view of the importance of FoxM1 for maintaining the capacity of cell proliferation, Yang et al. (38) confirmed that the down-regulation of FoxM1 expression could reduce the expression of cell cycle genes and diminish the proliferation and growth of breast cancer cells. Loss of FoxM1 expression also repressed proliferation without inducing the apoptosis of breast cancer cell lines independent of estrogen receptor (ER) status (39). A study by Hwang et al. showed that Moracin D repressed proliferation and induced apoptosis in breast cancer cells via attenuating the expression of FoxM1 related proteins and signaling pathways (40).
Additionally, emerging evidences have revealed that some microRNAs (miRNAs) are also implicated in the growth and proliferation of breast cancer cells by directly or indirectly regulating FoxM1 expression. The study by Zhang et al. (41) proved that DEP (dishevelled, EGL-10, pleckstrin) domain-containing 1 was negatively regulated by miR-26b, which its overexpression boosted tumor cells proliferation and growth via increasing the expression of FoxM1 in breast cancer. The experimental results of Yuan et al. (42) showed that overexpression of miR-802 could inhibit the proliferation of breast cancer cells in vitro as well as tumor growth in vivo through targeting the down-regulation of FoxM1 levels (Figure 3).
Roles of FoxM1 in breast cancer angiogenesis
Angiogenesis is a homeostatic process that not only occurs during embryogenesis, but also during the normal physiological repair processes as well as in various solid tumors, which is modulated by multiple factors and signaling pathways (43,44). Published research suggests that the occurrence and progression of malignant tumors mostly depend on the formation of new capillary blood vessels, which contribute fundamentally to supply the essential nutrients and oxygen, as well as for disposing metabolic wastes to promote tumor growth and metastasis (44). A growing body of evidence has shown that various growth factors and cytokines engage in regulating tumor angiogenesis, among which vascular endothelial growth factor (VEGF) and the VEGF receptor play the most vital role (45,46).
FoxM1, a crucial regulator of angiogenesis and carcinogenesis, plays a fundamental role in the modulation of VEGF transcription (47,48). Song et al. revealed that FoxM1 bound to the forkhead response element which presented on the VEGF gene promoter to facilitate VEGF transcription, thereby stimulating the formation of tumor angiogenesis (49). Ahmad et al. (50) used siRNA approach to down-regulate FoxM1 expression in MDA-MB-231 and SUM149 breast cancer cells with high FoxM1 expression. They found that as FoxM1 expression declined, VEGF activity was significantly decreased, and the capability of breast cancer cells to migrate and infiltrate was also attenuated.
It can be seen that FoxM1 can modulate the metastasis of tumor cells via regulating the activity of VEGF. A study by Zanin et al. (51) showed that FoxM1 was a crucial molecular partner of high mobility group A1, and further experimental results proved that their cooperative function could stimulate breast tumor angiogenic processes through in vitro and in vivo models. In another study, FoxM1 and transcription factor forkhead box protein O3 (FoxO3) in breast cancer cells competitively bound to the forkhead response element of the VEGF gene promoter (52). FoxM1 promoted the transcriptional expression of VEGF by binding to the promoter region of VEGF (53). In addition, it was reported that platelet-derived growth factor (PDGF) receptor signaling via the activation of PDGF and PDGF receptors was also closely correlated with tumor angiogenesis. A study by Yu et al. confirmed that elevated FoxM1 expression could up-regulate the expression of PDGF-A via binding to the PDGF-A promoter, thereby activating the V-akt murine thymoma viral oncogene homolog (Akt) pathway and promoting breast cancer cell growth and tumorigenesis (Figure 3) (54).
Of course, the entire process of human breast tumor formation and growth may contain more regulatory proteins or signaling pathways related to FoxM1.
Roles of FoxM1 in breast cancer invasion and metastasis
Tumor metastasis refers to a series of dynamic multi-stage processes covering local invasion of tumor cells around the original lesion, stimulation of angiogenesis and lymph angiogenesis, formation of micrometastasis, transportation of tumor cells, and survival, colonization, and growth at the secondary site (55,56). For most types of breast cancer, tumor metastasis is the prominent cause of cancer-related deaths (57,58). Generally, alterations of various molecules (including abnormal expression of miRNAs, the up-regulation of carcinogenes, the down-regulation of tumor suppressor genes, and a change in growth factors) are strongly correlated with complicated pathways and dynamic interactions in breast cancer, which play crucial roles in tumor metastatic processes (59,60). Furthermore, substantial evidence has confirmed that FoxM1 engages in each step of metastasis (from tumor occurrence to metastasis), demonstrating that FoxM1 is necessary for metastasis in breast cancer (61-63).
Cell invasiveness is enhanced, and the degradation of the basement membrane and the extracellular matrix (ECM) together give rise to the infiltration and metastasis of tumor cells (64). MMP, a collective name for a series of enzymes that are predominantly responsible for degrading various protein components of the ECM, can destroy the histological barrier of tumor cell invasion, and be intimately associated with tumor invasion and metastasis (65,66). Among them, the most important ones are MMP-2 and MMP-9, which degrade matrix membrane collagen. It has been strongly demonstrated that FoxM1 can directly or indirectly promote the expression of metalloproteinases such as MMP-2 and MMP-9 to enhance cell invasion and migration (67-69). Furthermore, previous studies have discovered that the down-regulation of FoxM1 levels can reverse the growth, migration, and invasiveness of breast cancer cells via reducing the expression of MMP-2, MMP-9, urokinase-type plasminogen activator (uPA), the uPA receptor, and VEGF (50,70).
In addition, a growing body of work has suggested that FoxM1 plays a central role in the invasive and migratory processes of malignant tumors by modulating EMT processes (71,72). EMT processes refer to when polarized epithelial cells lose their epithelial properties and acquire more mesenchymal phenotypes via cytoskeletal rearrangement and adhesion, thereby changing in cell structure and morphology, resulting in elevating the migratory and invasive capabilities of tumor cells (73,74). Some studies have demonstrated that EMT processes can be directly or indirectly regulated by FoxM1 to facilitate the invasion and migration of tumor cells (75-77). After the occurrence of EMT processes, tumor cells with epithelial characteristics show significantly elevated invasion and migration capabilities. Hence, EMT processes act as a necessary part of the invasion and migration of various tumors (78). Furthermore, it has been convincingly proven that the transformation of EMT processes promote the invasiveness and metastasis of breast cancer (79,80).
The accumulating evidence has revealed that various transcription factors (including FoxM1) affect the progression of EMT via diverse signaling pathways such as nuclear factor κB (NF-κB), transforming growth factor-β (TGF-β), Notch, hypoxia-inducible factor alpha, Wnt/β-catenin, the Fox family, and STAT3 (81-83). The activity and expression of FoxM1 can be regulated by numerous molecules and signaling pathways during the EMT processes of breast cancer. Yang et al. (84) confirmed that FoxM1-mediated EMT processes in breast cancer cells occurred in part through stimulating the expression of EMT-related transcription factors such as Slug, whereas inhibition of FoxM1 levels had the opposite effect. FoxM1 seemed to play a crucial role in EMT driven by constitutive cell surface receptor kinase signaling such as insulin-like growth factor-1 receptor (IGF-1R) signaling (85). Experimental results further showed that the enforced expression of FoxM1 facilitated the resistance to kinase targeting strategies, whereas its blockade suppress the EMT and invasion of kinase-driven breast cancer cells (85). The study of Xue et al. (86) confirmed that FoxM1 boosted the metastasis of breast cancer cells through activating the interaction between SMAD3 and the TGF-β pathway.
Recent studies showed that EMT processes could also be suppressed or promoted by non-coding RNAs (ncRNAs). Some miRNAs can also participate in EMT processes by regulating FoxM1 expression. For instance, the enforced expression of miR-671-5p gave rise to a shift from EMT to mesenchymal to epithelial transition phenotypes via directly down-regulating the expression of FoxM1, thus reducing the proliferation and invasion of MDA-MB-231 breast cancer cells (87). FoxM1 served as an hsa-miR-4756-3p target gene in TNBC, and blockade of FoxM1 totally suppressed hsa-miR-4756-3p-induced cell EMT, TGF-β1 signaling, migration and metastasis, which demonstrated that hsa-miR-4756-3p acted through the FoxM1-TGFβ1-EMT pathway (Figure 3) (88).
Generally speaking, the roles of FoxM1 in breast cancer invasion and metastasis maybe depend on its functions in modulating the expression of proteolysis-related genes and its contribution to EMT processes and angiogenesis. Forced expression of FoxM1 can induce EMT processes of breast cancer cells, whereas its blockade has the opposite influence, suggesting that FoxM1 can boost the infiltration and migration of breast cancer cells by regulating EMT processes.
Link between FoxM1 and ncRNAs in breast cancer
Supporting previously published research displayed that ncRNAs are the non-protein-coding RNAs, which are generally produced through eukaryotic genomes, and include miRNAs, long non-coding RNAs (lncRNAs), and circular RNAs. Approximately 99% of the human genomes lack protein-coding function, but published studies have indicated that ncRNAs are correlated with a wide spectrum of physiological processes, such as RNA splicing and translation, epigenetic regulation, and DNA replication (89). Moreover, various studies have also proven that ncRNAs play a crucial role in regulating genesis and development of many human tumors (90,91). In this review, we will next discuss the connection between FoxM1 and ncRNAs in breast cancer.
Speaking of miRNAs, it has been proved that miRNAs are a type of endogenous, small, single-stranded ncRNAs with a length of about 18–24 bases, which regulate the expression of genes at the post-transcriptional level (92,93). Numerous studies have indicated that miRNAs play predominant roles in the process of tumor cell proliferation, invasion, metastasis, relapse, and drug resistance (2,94,95). Mounting evidence has proven that various miRNAs, which abnormally express in breast cancer, engage in repressing or facilitating the genesis and progression of breast cancer via targeting the down-regulation of FoxM1 expression, such as miR-802, miR-4756-3p, miR-34a, miR-671-5p, and miR-23a (as shown in Table 1).
Table 1
| miRNA expression | Effect | First author/year | References |
|---|---|---|---|
| miR-802 ↓ down | Down-regulate protein expression levels of FoxM1 | Yuan/2015 | (42) |
| miR-4756-3p ↓ down | Gu/2019 | (88) | |
| miR-23a ↑ up | Eissa/2015 | (96) | |
| miR-34a ↓ down | Bayraktar/2018 | (97) | |
| miR-671-5p ↓ down | Tan/2019 | (98) |
FoxM1, forkhead box transcription factor M1.
In addition, some recent studies have revealed that lncRNAs, with a size longer than 200 nucleotides in the transcript, also play crucial roles in regulating the initiation and development of many tumors (99-101). Several published studies have indicated that lncRNAs can affect FoxM1 expression by directly acting on pivotal molecules or by functioning as a “sponge” for miRNAs, which further are conducive to the progression of breast cancer (as shown in Table 2). Milevskiy et al. (102) discovered that the expression of lncRNA-HOTAIR was positively correlated with FoxM1 protein expression. The authors further found that HOTAIR was co-expressed with FoxA1 and FoxM1 in human epidermal growth factor receptor 2 (HER2)-enriched tumors, and these factors strengthened the prognostic capability of HOTAIR in aggressive HER2 positive breast cancer.
Table 2
| lncRNA expression | Effect | First author/year | References |
|---|---|---|---|
| HOTAIR ↑ up | Transcriptionally up-regulated by FoxM1 proteins | Milevskiy/2016 | (102) |
| FBXL19-AS1 ↑ up | Up-regulate FoxM1 via absorbing miR-876-5p | Dong/2019 | (103) |
| LINC00885 ↑ up | Activate the EREG, EGFR, and FoxM1 pathways | Abba/2020 | (104) |
FoxM1, forkhead box transcription factor M1; EREG, epiregulin; EGFR, epidermal growth factor receptor.
To sum up, present relevant reports have not yet fully clarified the mechanism of action between FoxM1 and ncRNAs in the occurrence and development of breast cancer. Consequently, further exploring the connection between FoxM1 and ncRNAs in breast cancer will facilitate a better understanding of the pathogenesis of breast cancer and provide new ideas for the treatment and prognosis of breast cancer patients.
Roles of FoxM1 in the drug resistance of breast cancer
Chemotherapy is currently a common method for the conventional and essential treatment of various tumors in the clinic to reduce the death of cancer patients (74,105). So far, there are numerous anticancer drugs used in the clinic, and therapy failure is usually due to drug resistance, which is the main hurdle to the successful treatment of diverse cancers (106). Drug resistance is divided into 2 major categories: inherent resistance or acquired resistance (53). There are diverse molecular mechanisms of cancer drug resistance including drug absorption and transport, rates of drug efflux, alterations in drug metabolism, mutation of drug targets, epigenetic changes, tumor related genes, tumor stemness, DNA repair, and tumor environment (107,108).
Meanwhile, the resistance of chemotherapeutic agents constitutes also a major obstacle for the effective treatment of breast cancer patients (109-111). Furthermore, substantial evidence has explained that FoxM1 is closely related to the drug resistance of diverse human tumors (gastric cancer, prostate cancer, and colorectal cancer) (112-115). Many studies have confirmed that when the level of FoxM1 expression is enhanced, it can cause the resistance of cancer cells to chemotherapy drugs, suggesting that FoxM1 plays a crucial role in the drug resistance of various tumors (116,117).
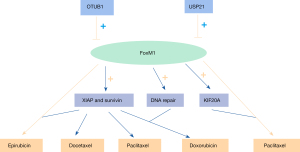
Karunarathna et al. (118) revealed that FoxM1 up-regulation was involved in genotoxic drug resistance in breast cancer. The authors discovered that OTU domain-containing ubiquitin aldehyde-binding protein 1 (OTUB1) positively modulated FoxM1 expression. In addition, experimental results indicated that because OTUB1 had little effect on FoxM1-deficient cells, targeting FoxM1 improved the proliferation rate and epirubicin resistance. These data indicated that OTUB1 restricted the ubiquitination and degradation of FoxM1 and was closely correlated with drug resistance in breast cancer. Nestal de Moraes et al. (119) demonstrated that the enforced expression of of FoxM1 strengthened the anti-apoptotic genes XIAP and survivin by interacting with their promoter regions, contributing to the chemoresistance of breast cancer cells to docetaxel, doxorubicin (Dox), epirubicin, and paclitaxel. Furthermore, co-expression of FoxM1, survivin, and nuclear XIAP were correlated with the poor prognoses of females with stage III breast invasive ductal carcinoma with markedly reduced 5 and 10 years overall survival rates versus females with tumors without these characteristics. Depletion of ubiquitin-specific protease 21 (USP21) down-regulated the expression of FoxM1 and obviously delayed cell cycle progression, resulting in the sensitization of basal-like breast cancer cell lines and mouse xenograft tumors to paclitaxel (120).
The overexpression of FoxM1 strengthened epirubicin-induced DNA damage repair and resistance to epirubicin in breast cancer cells (121). A study by Park et al. (122) found that silencing of FoxM1 expression strengthened the sensitivity of breast cancer cells to Dox via directly down-regulating the expression of DNA repair genes. The study by Khongkow et al. (123) found that FoxM1 depletion resensitized MCF-7 breast cancer cells to epirubicin-induced cellular senescence. Khongkow et al. (124) also proposed that paclitaxel resistance might be mediated by FoxM1 through enhancing the activity of the promoter for the transcriptional activity of KIF20A. FoxM1 and KIF20A played pivotal roles in the formation of normal mitotic spindles, thereby interfering with the activity of paclitaxel.
In addition, more and more research articles have shown that FoxM1 also participates in the regulation of autophagy, tumor stem cell formation, and cell senescence to promote the genesis and development of breast tumors. For instance, blockade of FoxM1 levels suppressed starvation and rapamycin-induced autophagy through modulating the transcriptional activity of the major autophagy regulators LC3 and Beclin-1 in human TNBC cells (125). FoxM1, via recruiting nuclear Aurora kinase A, engaged in a tightly coupled positive feedback loop to boost the breast cancer stem cell (BCSC) phenotype, thus strengthening the tumorigenicity and self-renewal ability of BCSCs (126). Up-regulation of FoxM1 levels was linked to an expansion of the cancer stem-like cell population, resulting in cell aggressiveness and resistance to endocrine therapies (127). FoxM1 also mediated the levels of NBS1 to repress cell senescence induced by DNA damage in breast cancer, while evading cellular senescence facilitated infinite replicative capacity to induce tumor development (123).
These findings have uncovered that FoxM1 is closely implicated in the drug resistance of breast cancer (Figure 4). An in-depth understanding of the underlying mechanisms of FoxM1 in breast cancer drug resistance is of great significance for the development of predictive biomarkers and novel chemotherapeutic strategies for drug resistance.
FoxM1 may act as a prognostic marker in breast cancer patients
FoxM1 is demonstrated to play significant roles in initiation and progression of breast cancer, including being closely related to tumor proliferation, angiogenesis, metastasis, invasion, and chemotherapeutic drug resistance. FoxM1 is aberrantly overexpressed in a variety of tumors, but is not expressed and undetectable in most normal mature tissue cells (128). Furthermore, a growing body of data has also indicated that the expression of FoxM1 is increased in breast cancer tissues, and is closely correlated with unfavorable clinical outcomes in patients (127). The results of these preclinical studies have shown that this transcription factor can be used as a favorable prognostic biomarker for breast cancer patients (129).
A study by Bektas et al. (130) reported that elevated protein levels of FoxM1 were linked to the adverse prognosis of breast cancer. Higher expression of FoxM1 in the tissues of patients with advanced stage (stage III and IV) Middle Eastern breast cancer indicated that this transcription factor could be used as an independent poor prognostic marker for this malignant tumor (131). In addition, Ahn et al. (132) demonstrated that increased expression of FoxM1 was associated with unfavorable clinicopathological characteristics such as lymphovascular invasiveness, advanced tumor stage, larger tumor size, and lymph node metastasis in ER-positive breast cancers by immunohistochemical staining on tissue microarray sections from 236 breast cancer patients. Furthermore, this study also showed that the up-regulation of FoxM1 expression was closely linked to aggressive phenotypes, poor prognosis of ER-positive breast cancer, as well as poor disease-free survival (DFS) and overall survival for patients. In another study, Abdeljaoued et al. (133) showed that enforced expression of FoxM1 was associated with higher histological grade, tumor stage, and Ki-67 proliferation index in male breast cancer (MBC). Moreover, high expression of FoxM1 in MBC patients was also significantly related to chemotherapy and endocrine resistance and shorter DFS. Taken together, FoxM1 can be used as a prognostic biomarker for breast cancer.
Roles of FoxM1 in therapeutics for breast cancer
Considering all the aforementioned studies, it is no surprise that some studies have pointed out that FoxM1 may be a promising prospective tumor-specific therapeutic target in breast cancer (134,135). Herein, we will summarize some of the breast cancer therapeutic strategies directly or indirectly targeting FoxM1 (as shown in Table 3).
Table 3
| Inhibitors/drugs | Description | Function | References |
|---|---|---|---|
| Imipramine blue | A new analogue of the antidepressant imipramine | Block the activity of FoxM1 and FoxM1-associated signaling to suppress breast cancer growth and metastasis | (136) |
| Thiostrepton | Thiazole antibiotic | Induce apoptosis and suppress the growth of breast cancer cells by down-regulating the expression of FoxM1 and cyclin B1 | (137) |
| Moracin D | 2-arylbenzofuran flavonoid | Inhibit the Wnt3a/FoxM1/β-catenin signaling pathway to induce apoptosis and suppress the proliferation of breast cancer | (40) |
| Panepoxydone | A type of NF-κB inhibitor | Down-regulate FoxM1 and reverse EMT in breast cancer | (138) |
| Sepin-1 | A potent non-competitive inhibitor of separase | Repress the expression of cell cycle driving genes and cell growth via down-regulating the expression of Raf and FoxM1 in breast cancer | (139) |
| Maslinic acid | A natural triterpene from Olea europaea L. | Suppress the MELK-FoxM1-ABCB1 signaling cascade to enhance docetaxel response in TNBC | (140) |
| 3,3'-diindolylmethane | A nontoxic dietary chemopreventive agent | (I) Down-regulate the expression of FoxM1 and its target genes and enhance the therapeutic efficacy of Taxotere in breast cancer | (141) |
| (II) Enhance the efficacy of Herceptin in breast cancer cells, accompanied by reducing the expression of FoxM1 | (142) | ||
| Casticin | An active ingredient extracted from the Fructus Viticis of traditional Chinese medicine | Reduce the expression of FoxM1 and induce the apoptosis of breast cancer cells via strengthening dephosphorylation of FOXO3a | (143) |
| Ursolic acid | 3β-hydroxy-12-urs-12-en-28-oic acid | Suppress the expression of cyclin D1/CDK4 and FoxM1, thus increasing the apoptosis of breast cancer cells | (144) |
| Lapatinib | HER2 inhibitor | Reduce FoxM1 expression at the protein, mRNA, and gene promoter levels in breast cancer sensitive cell lines to block the progression of tumor cells | (145) |
| FDI-6 | 3-amino-N-(4-fluorophenyl)-6-(thiophen-2-yl)-4-(trifluoromethyl)thieno[2,3-b]pyridine-2-carboxamide TFA | (I) Repress the binding of FoxM1 to target DNA and block the downstream transcriptional activation of FoxM1-controlled genes in cells | (146) |
| (II) Suppress cell growth and increase the apoptosis of TNBC cells via down-regulating FoxM1 levels and its pivotal oncogenic targets, including cyclin B1, Snail, and Slug | (147) | ||
| TFI10 | Modified thiazolidinedione | Decrease the mRNA levels of FoxM1 target genes | (148) |
| MG132 | Proteasome inhibitor | Promote tumor cell apoptosis via inhibiting FoxM1 transcriptional activity and FoxM1 expression | (149) |
| Honokiol | Anti-inflammatory, anti-oxidant | Suppress FoxM1-mediated transcription and FoxM1 protein expression | (150) |
| Morin | A flavonoid extracted from the Moraceace family | Maintain cell cycle arrest via activating ERK and repressing FoxM1 signaling pathways to induce p21 expression | (151) |
| Apigenin | A flavone found in several plant foods | Repress the Akt/FoxM1 signaling pathway via reducing the expression of FoxM1 | (39) |
FoxM1, forkhead box transcription factor M1; NF-κB, nuclear factor κB; EMT, epithelial-mesenchymal transition; MELK, maternal embryonic leucine-zipper kinase; TNBC, triple-negative breast cancer; FOXO3a, transcription factor forkhead box protein O3; CDK4, cyclin dependent kinase 4; HER2, human epidermal growth factor receptor 2; ERK, extracellular signal-regulated kinase; Akt, V-akt murine thymoma viral oncogene homolog.
In addition, several current studies have found that some novel compounds and proteins can suppress further development of breast cancer cells via eliminating the function of FoxM1. Ye et al. (152) identified the three new downstream target proteins (ACSL4, CGGBP1, and PGRMC2) of FoxM1 in breast cancer MDA-MB-231 cells by quantitative proteomic analysis. Further functional experiments also verified that depletion of the three proteins obviously retarded the capability of MDA-MB-231 cell migration, which was consistent with the phenotype of FoxM1 knockdown. The above results uncover that new potential downstream effectors of FoxM1 may act as novel favorable therapeutic targets in breast cancer. Another study by Ziegler et al. (153) demonstrated that these novel compounds (including 1,1-diarylethylene mono, diamines, and their corresponding methiodide salts) suppressed FoxM1 activity and breast tumor cell proliferation and growth via cell cycle arrest, and induced apoptosis. Furthermore, Dey et al. (154) revealed that the compounds NB-73 and NB-115, through decreasing the expression level of FoxM1 target genes, attenuated the growth, invasiveness, distant metastasis, and the expression of important proteins associated with EMT in TNBC cells.
On the other hand, the combination therapy of Dox and liposomal FoxM1 aptamer (Lip-FoxM1apt) strikingly enhanced both the cytotoxicity of Dox in breast cancer cells as well as the apoptosis induced by Dox (155). In a mouse model, the use of Lip-FoxM1apt improved the anti-tumor efficacy of Dox, which was markedly more effective than Dox monotherapy. Another study (156) also indicated that liposome-encapsulated thiostrepton (TSLP) displayed a higher potential in reducing FoxM1 expression in MCF-7 cells than free thiostrepton, and further experimental results revealed that TSLP significantly elevated the effectiveness and specificity of thiostrepton in decreasing the cell viability of MCF-7.
The above evidence suggests that FoxM1 may prove to be a novel potential therapeutic target of breast cancer, which may help suppress tumor cell development and strengthen the response of breast cancer cells to drug treatment.
Conclusions
Overall, FoxM1 plays indispensable roles in the occurrence, development, and drug resistance of breast cancer. Combined with the current research, a common conclusion can be drawn, that is, targeting the silencing of FoxM1 can suppress the growth, proliferation, invasion, and metastasis of breast cancer (20,49). Therefore, FoxM1 may act as a powerful biomarker in the therapy and prognosis of breast cancer, and may become a novel potential therapeutic target for breast cancer in the clinic (157).
However, the biological functions and molecular mechanism of FoxM1 in breast cancer have remained largely unknown, and it is still a challenge to develop drugs with specific functions as FoxM1 inhibitors. Further, little is known about isoforms of FoxM1 in different types of breast cancer, which may become a predominant theme for the future researches. Future research should pay more attention to study the functions and anti-tumor activity of isoform-specific FoxM1 inhibitors in different subtypes of breast cancer. In addition, researchers should clarify which isoforms are most likely to benefit from treatments targeting FoxM1, particularly in drug-resistant and metastatic breast cancers (158). Additionally, some studies have shown that certain ncRNAs can directly or indirectly regulate the expression of FoxM1, which may provide a promising therapeutic intervention target for breast cancer treatment in the future (159,160).
In recent years, the understanding of the tumor microenvironment of different subtypes of breast cancer has been continuously improved, and breast cancer has been considered as an immunogenic tumor. At the same time, new researches show that immunotherapy, as a new auxiliary method for the treatment of breast cancer, has achieved good clinical results, especially in TNBC (161-163). At present, exploring tumor immune response and identifying biomarkers that can predict immunotherapy have become a major trend, and several studies have reported that FoxM1 was linked to immunotherapy in certain human cancers (164). A study by Zhang et al. (165) found that expression levels of Janus kinase 2 and FoxM1 were related to the immune infiltration in non-small cell lung cancer, and their high expression could independently predict the clinical outcome of lung squamous cell carcinoma patients receiving immune checkpoint inhibitors. Su et al. (166) demonstrated that the immunity induced via dendritic cells loaded with cytoplasmic transduction peptide (CTP)-FoxM1 could meaningfully repress tumor growth and metastasis in hepatocellular carcinoma-bearing mice, which was more effective than the immunity induced through dendritic cells loaded with FoxM1 or CTP alone. Cyclin D1 and cyclin-dependent kinase (CDK) inhibitors could block the phosphorylation process of retinoblastoma proteins (including FoxM1), change the tumor cells cycle, and induce anti-tumor immune activity in breast cancer (167). These results indicated that FoxM1 may be a key molecule for modulating immune response through CDK inhibitors in the treatment of breast cancer (167). However, there are few reports about the effect of FoxM1 on breast cancer immunotherapy, which can be used as a pivotal research direction in the future.
Eventually, related articles point out that there are four subtypes of FoxM1 due to the different splicing of exons Va and VIIa (15), so it is of great significance to determine the interaction among subtypes and whether there is isoforms conversion among FoxM1a and/or FoxM1b and/or FoxM1c and/or FoxM1d on the occurrence and development of breast cancer. Further investigations that clarify the epigenetic mechanism of the differential expression of FoxM1 subtypes in breast cancer cells are equally important.
Acknowledgments
Funding: This work was supported by the Xinglin Scholar Research Premotion Project of Chengdu University of Traditional Chinese Medicine (grant No. ZRYY2023), and the Sichuan Collaborative Innovation Center of Elderly Care and Health (grant No. 19Z15).
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-5271
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-5271). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhang X, Zhang L, Du Y, et al. A novel FOXM1 isoform, FOXM1D, promotes epithelial-mesenchymal transition and metastasis through ROCKs activation in colorectal cancer. Oncogene 2017;36:807-19. [Crossref] [PubMed]
- Li X, Zeng Z, Wang J, et al. MicroRNA-9 and breast cancer. Biomed Pharmacother 2020;122:109687 [Crossref] [PubMed]
- Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 2019;144:1941-53. [Crossref] [PubMed]
- Fidler MM, Bray F, Soerjomataram I. The global cancer burden and human development: A review. Scand J Public Health 2018;46:27-36. [Crossref] [PubMed]
- Gu G, Dustin D, Fuqua SA. Targeted therapy for breast cancer and molecular mechanisms of resistance to treatment. Curr Opin Pharmacol 2016;31:97-103. [Crossref] [PubMed]
- Barger CJ, Branick C, Chee L, et al. Pan-Cancer Analyses Reveal Genomic Features of FOXM1 Overexpression in Cancer. Cancers (Basel) 2019;11:251. [Crossref] [PubMed]
- Laissue P. The forkhead-box family of transcription factors: key molecular players in colorectal cancer pathogenesis. Mol Cancer 2019;18:5. [Crossref] [PubMed]
- Wierstra I. The transcription factor FOXM1 (Forkhead box M1): proliferation-specific expression, transcription factor function, target genes, mouse models, and normal biological roles. Adv Cancer Res 2013;118:97-398. [Crossref] [PubMed]
- Korver W, Roose J, Heinen K, et al. The human TRIDENT/HFH-11/FKHL16 gene: structure, localization, and promoter characterization. Genomics 1997;46:435-42. [Crossref] [PubMed]
- Kalathil D, John S, Nair AS. FOXM1 and Cancer: Faulty Cellular Signaling Derails Homeostasis. Front Oncol 2020;10:626836 [Crossref] [PubMed]
- Zhou Y, Wang Q, Chu L, et al. FOXM1c promotes oesophageal cancer metastasis by transcriptionally regulating IRF1 expression. Cell Prolif 2019;52:e12553 [Crossref] [PubMed]
- Klinhom-On N, Seubwai W, Sawanyawisuth K, et al. FOXM1c is the predominant FOXM1 isoform expressed in cholangiocarcinoma that associated with metastatic potential and poor prognosis of patients. Heliyon 2021;7:e06846 [Crossref] [PubMed]
- Zhang W, Zhang X, Huang S, et al. FOXM1D potentiates PKM2-mediated tumor glycolysis and angiogenesis. Mol Oncol 2021;15:1466-85. [Crossref] [PubMed]
- Wang Q, Zhang P, Zhang W, et al. PI3K activation is enhanced by FOXM1D binding to p110 and p85 subunits. Signal Transduct Target Ther 2020;5:105. [Crossref] [PubMed]
- Lam AK, Ngan AW, Leung MH, et al. FOXM1b, which is present at elevated levels in cancer cells, has a greater transforming potential than FOXM1c. Front Oncol 2013;3:11. [PubMed]
- Wang CM, Liu R, Wang L, et al. SUMOylation of FOXM1B alters its transcriptional activity on regulation of MiR-200 family and JNK1 in MCF7 human breast cancer cells. Int J Mol Sci 2014;15:10233-51. [Crossref] [PubMed]
- Wierstra I. FOXM1 (Forkhead box M1) in tumorigenesis: overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Adv Cancer Res 2013;119:191-419. [Crossref] [PubMed]
- Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer 2007;7:847-59. [Crossref] [PubMed]
- Hong H, Zhu H, Zhao S, et al. The novel circCLK3/miR-320a/FoxM1 axis promotes cervical cancer progression. Cell Death Dis 2019;10:950. [Crossref] [PubMed]
- Saba R, Alsayed A, Zacny JP, et al. The Role of Forkhead Box Protein M1 in Breast Cancer Progression and Resistance to Therapy. Int J Breast Cancer 2016;2016:9768183 [Crossref] [PubMed]
- Zhang Z, Tu K, Liu F, et al. FoxM1 promotes the migration of ovarian cancer cell through KRT5 and KRT7. Gene 2020;757:144947 [Crossref] [PubMed]
- Xu MD, Wang Y, Weng W, et al. A Positive Feedback Loop of lncRNA-PVT1 and FOXM1 Facilitates Gastric Cancer Growth and Invasion. Clin Cancer Res 2017;23:2071-80. [Crossref] [PubMed]
- Liu C, Shi J, Li Q, et al. STAT1-mediated inhibition of FOXM1 enhances gemcitabine sensitivity in pancreatic cancer. Clin Sci (Lond) 2019;133:645-63. [Crossref] [PubMed]
- Yang Y, Jiang H, Li W, et al. FOXM1/DVL2/Snail axis drives metastasis and chemoresistance of colorectal cancer. Aging (Albany NY) 2020;12:24424-40. [Crossref] [PubMed]
- Liao GB, Li XZ, Zeng S, et al. Regulation of the master regulator FOXM1 in cancer. Cell Commun Signal 2018;16:57. [Crossref] [PubMed]
- Yue M, Li S, Yan G, et al. Paeoniflorin inhibits cell growth and induces cell cycle arrest through inhibition of FoxM1 in colorectal cancer cells. Cell Cycle 2018;17:240-9. [Crossref] [PubMed]
- Kelleher FC, O'Sullivan H. FOXM1 in sarcoma: role in cell cycle, pluripotency genes and stem cell pathways. Oncotarget 2016;7:42792-804. [Crossref] [PubMed]
- Wierstra I, Alves J. FOXM1, a typical proliferation-associated transcription factor. Biol Chem 2007;388:1257-74. [Crossref] [PubMed]
- Wu CH, Yeh CT, Lin KH. Thyroid hormones suppress FOXM1 expression to reduce liver cancer progression. Oncol Rep 2020;44:1686-98. [Crossref] [PubMed]
- Hamurcu Z, Ashour A, Kahraman N, et al. FOXM1 regulates expression of eukaryotic elongation factor 2 kinase and promotes proliferation, invasion and tumorgenesis of human triple negative breast cancer cells. Oncotarget 2016;7:16619-35. [Crossref] [PubMed]
- Mancini M, De Santis S, Monaldi C, et al. Hyper-activation of Aurora kinase a-polo-like kinase 1-FOXM1 axis promotes chronic myeloid leukemia resistance to tyrosine kinase inhibitors. J Exp Clin Cancer Res 2019;38:216. [Crossref] [PubMed]
- Murakami H, Aiba H, Nakanishi M, et al. Regulation of yeast forkhead transcription factors and FoxM1 by cyclin-dependent and polo-like kinases. Cell Cycle 2010;9:3233-42. [Crossref] [PubMed]
- Wang X, Quail E, Hung NJ, et al. Increased levels of forkhead box M1B transcription factor in transgenic mouse hepatocytes prevent age-related proliferation defects in regenerating liver. Proc Natl Acad Sci U S A 2001;98:11468-73. [Crossref] [PubMed]
- Yang N, Wang C, Wang J, et al. Aurora kinase A stabilizes FOXM1 to enhance paclitaxel resistance in triple-negative breast cancer. J Cell Mol Med 2019;23:6442-53. [Crossref] [PubMed]
- Zhang J, Lu J, Chen Y, et al. WHSC1 promotes wnt/β-catenin signaling in a FoxM1-dependent manner facilitating proliferation, invasion and epithelial-mesenchymal transition in breast cancer. J Recept Signal Transduct Res 2020;40:410-8. [Crossref] [PubMed]
- Hu G, Yan Z, Zhang C, et al. FOXM1 promotes hepatocellular carcinoma progression by regulating KIF4A expression. J Exp Clin Cancer Res 2019;38:188. [Crossref] [PubMed]
- Pan H, Zhu Y, Wei W, et al. Transcription factor FoxM1 is the downstream target of c-Myc and contributes to the development of prostate cancer. World J Surg Oncol 2018;16:59. [Crossref] [PubMed]
- Yang C, Chen H, Yu L, et al. Inhibition of FOXM1 transcription factor suppresses cell proliferation and tumor growth of breast cancer. Cancer Gene Ther 2013;20:117-24. [Crossref] [PubMed]
- Pham TH, Page YL, Percevault F, et al. Apigenin, a Partial Antagonist of the Estrogen Receptor (ER), Inhibits ER-Positive Breast Cancer Cell Proliferation through Akt/FOXM1 Signaling. Int J Mol Sci 2021;22:470. [Crossref] [PubMed]
- Hwang SM, Lee HJ, Jung JH, et al. Inhibition of Wnt3a/FOXM1/β-Catenin Axis and Activation of GSK3β and Caspases are Critically Involved in Apoptotic Effect of Moracin D in Breast Cancers. Int J Mol Sci 2018;19:2681. [Crossref]
- Zhang L, Du Y, Xu S, et al. DEPDC1, negatively regulated by miR-26b, facilitates cell proliferation via the up-regulation of FOXM1 expression in TNBC. Cancer Lett 2019;442:242-51. [Crossref] [PubMed]
- Yuan F, Wang W. MicroRNA-802 suppresses breast cancer proliferation through downregulation of FoxM1. Mol Med Rep 2015;12:4647-51. [Crossref] [PubMed]
- Li S, Xu HX, Wu CT, et al. Angiogenesis in pancreatic cancer: current research status and clinical implications. Angiogenesis 2019;22:15-36. [Crossref] [PubMed]
- Viallard C, Larrivée B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis 2017;20:409-26. [Crossref] [PubMed]
- Chen Y, Zhang L, Liu WX, et al. VEGF and SEMA4D have synergistic effects on the promotion of angiogenesis in epithelial ovarian cancer. Cell Mol Biol Lett 2018;23:2. [Crossref] [PubMed]
- Waldner MJ, Neurath MF. Targeting the VEGF signaling pathway in cancer therapy. Expert Opin Ther Targets 2012;16:5-13. [Crossref] [PubMed]
- Yang X, Shi Y, Yan J, et al. Downregulation of FoxM1 inhibits cell growth and migration and invasion in bladder cancer cells. Am J Transl Res 2018;10:629-38. [PubMed]
- Zhang HG, Xu XW, Shi XP, et al. Overexpression of forkhead box protein M1 (FOXM1) plays a critical role in colorectal cancer. Clin Transl Oncol 2016;18:527-32. [Crossref] [PubMed]
- Song X, Fiati Kenston SS, Zhao J, et al. Roles of FoxM1 in cell regulation and breast cancer targeting therapy. Med Oncol 2017;34:41. [Crossref] [PubMed]
- Ahmad A, Wang Z, Kong D, et al. FoxM1 down-regulation leads to inhibition of proliferation, migration and invasion of breast cancer cells through the modulation of extra-cellular matrix degrading factors. Breast Cancer Res Treat 2010;122:337-46. [Crossref] [PubMed]
- Zanin R, Pegoraro S, Ros G, et al. HMGA1 promotes breast cancer angiogenesis supporting the stability, nuclear localization and transcriptional activity of FOXM1. J Exp Clin Cancer Res 2019;38:313. [Crossref] [PubMed]
- Karadedou CT, Gomes AR, Chen J, et al. FOXO3a represses VEGF expression through FOXM1-dependent and -independent mechanisms in breast cancer. Oncogene 2012;31:1845-58. [Crossref] [PubMed]
- Yao S, Fan LY, Lam EW. The FOXO3-FOXM1 axis: A key cancer drug target and a modulator of cancer drug resistance. Semin Cancer Biol 2018;50:77-89. [Crossref] [PubMed]
- Yu G, Zhou A, Xue J, et al. FoxM1 promotes breast tumorigenesis by activating PDGF-A and forming a positive feedback loop with the PDGF/AKT signaling pathway. Oncotarget 2015;6:11281-94. [Crossref] [PubMed]
- Suhail Y, Cain MP, Vanaja K, et al. Systems Biology of Cancer Metastasis. Cell Syst 2019;9:109-27. [Crossref] [PubMed]
- Stoletov K, Beatty PH, Lewis JD. Novel therapeutic targets for cancer metastasis. Expert Rev Anticancer Ther 2020;20:97-109. [Crossref] [PubMed]
- Liang Y, Zhang H, Song X, et al. Metastatic heterogeneity of breast cancer: Molecular mechanism and potential therapeutic targets. Semin Cancer Biol 2020;60:14-27. [Crossref] [PubMed]
- Jazieh K, Bell R, Agarwal N, et al. Novel targeted therapies for metastatic breast cancer. Ann Transl Med 2020;8:907. [Crossref] [PubMed]
- Scully OJ, Bay BH, Yip G, et al. Breast cancer metastasis. Cancer Genomics Proteomics 2012;9:311-20. [PubMed]
- Sharma R, Sharma R, Khaket TP, et al. Breast cancer metastasis: Putative therapeutic role of vascular cell adhesion molecule-1. Cell Oncol (Dordr) 2017;40:199-208. [Crossref] [PubMed]
- Raychaudhuri P, Park HJ. FoxM1: a master regulator of tumor metastasis. Cancer Res 2011;71:4329-33. [Crossref] [PubMed]
- Yau C, Meyer L, Benz S, et al. FOXM1 cistrome predicts breast cancer metastatic outcome better than FOXM1 expression levels or tumor proliferation index. Breast Cancer Res Treat 2015;154:23-32. [Crossref] [PubMed]
- Ferrer CM, Lu TY, Bacigalupa ZA, et al. O-GlcNAcylation regulates breast cancer metastasis via SIRT1 modulation of FOXM1 pathway. Oncogene 2017;36:559-69. [Crossref] [PubMed]
- Walma DAC, Yamada KM. The extracellular matrix in development. Development 2020;147:dev175596 [Crossref] [PubMed]
- Claesson-Welsh L. How the matrix metalloproteinase MMP14 contributes to the progression of colorectal cancer. J Clin Invest 2020;130:1093-5. [Crossref] [PubMed]
- Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 2010;141:52-67. [Crossref] [PubMed]
- Zhu X, Yu M, Wang K, et al. FoxM1 affects adhesive, migratory, and invasive abilities of human retinoblastoma Y-79 cells by targeting matrix metalloproteinase 2. Acta Biochim Biophys Sin (Shanghai) 2020;52:294-301. [Crossref] [PubMed]
- Ahmed M, Uddin S, Hussain AR, et al. FoxM1 and its association with matrix metalloproteinases (MMP) signaling pathway in papillary thyroid carcinoma. J Clin Endocrinol Metab 2012;97:E1-E13. [Crossref] [PubMed]
- He SY, Shen HW, Xu L, et al. FOXM1 promotes tumor cell invasion and correlates with poor prognosis in early-stage cervical cancer. Gynecol Oncol 2012;127:601-10. [Crossref] [PubMed]
- Ahmad A, Wang Z, Kong D, et al. Retraction Note to: FoxM1 down-regulation leads to inhibition of proliferation, migration and invasion of breast cancer cells through the modulation of extra-cellular matrix degrading factors. Breast Cancer Res Treat 2016;158:607. [Crossref] [PubMed]
- Wang Y, Yao B, Wang Y, et al. Increased FoxM1 expression is a target for metformin in the suppression of EMT in prostate cancer. Int J Mol Med 2014;33:1514-22. [Crossref] [PubMed]
- Meng FD, Wei JC, Qu K, et al. FoxM1 overexpression promotes epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma. World J Gastroenterol 2015;21:196-213. [Crossref] [PubMed]
- Tang L, Chen Y, Chen H, et al. DCST1-AS1 Promotes TGF-β-Induced Epithelial-Mesenchymal Transition and Enhances Chemoresistance in Triple-Negative Breast Cancer Cells via ANXA1. Front Oncol 2020;10:280. [Crossref] [PubMed]
- Du B, Shim JS. Targeting Epithelial-Mesenchymal Transition (EMT) to Overcome Drug Resistance in Cancer. Molecules 2016;21:965. [Crossref] [PubMed]
- Zhang J, Chen XY, Huang KJ, et al. Expression of FoxM1 and the EMT-associated protein E-cadherin in gastric cancer and its clinical significance. Oncol Lett 2016;12:2445-50. [Crossref] [PubMed]
- Fei BY, He X, Ma J, et al. FoxM1 is associated with metastasis in colorectal cancer through induction of the epithelial-mesenchymal transition. Oncol Lett 2017;14:6553-61. [Crossref] [PubMed]
- Kong FF, Qu ZQ, Yuan HH, et al. Overexpression of FOXM1 is associated with EMT and is a predictor of poor prognosis in non-small cell lung cancer. Oncol Rep 2014;31:2660-8. [Crossref] [PubMed]
- Zhao M, Ang L, Huang J, et al. MicroRNAs regulate the epithelial-mesenchymal transition and influence breast cancer invasion and metastasis. Tumour Biol 2017;39:1010428317691682 [Crossref] [PubMed]
- Fenizia C, Bottino C, Corbetta S, et al. SMYD3 promotes the epithelial-mesenchymal transition in breast cancer. Nucleic Acids Res 2019;47:1278-93. [Crossref] [PubMed]
- Hong D, Fritz AJ, Zaidi SK, et al. Epithelial-to-mesenchymal transition and cancer stem cells contribute to breast cancer heterogeneity. J Cell Physiol 2018;233:9136-44. [Crossref] [PubMed]
- Zhang J, Xu Z, Dai H, et al. Silencing of Forkhead Box M1 Reverses Transforming Growth Factor-beta1-Induced Invasion and Epithelial-Mesenchymal Transition of Endometriotic Epithelial Cells. Gynecol Obstet Invest 2019;84:485-94. [Crossref] [PubMed]
- Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: yes, no, maybe? Curr Opin Cell Biol 2016;43:7-13. [Crossref] [PubMed]
- Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol 2017;11:28-39. [Crossref] [PubMed]
- Yang C, Chen H, Tan G, et al. FOXM1 promotes the epithelial to mesenchymal transition by stimulating the transcription of Slug in human breast cancer. Cancer Lett 2013;340:104-12. [Crossref] [PubMed]
- Peake BF, Eze SM, Yang L, et al. Growth differentiation factor 15 mediates epithelial mesenchymal transition and invasion of breast cancers through IGF-1R-FoxM1 signaling. Oncotarget 2017;8:94393-406. [Crossref] [PubMed]
- Xue J, Lin X, Chiu WT, et al. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-β-dependent cancer metastasis. J Clin Invest 2014;124:564-79. [Crossref] [PubMed]
- Tan X, Fu Y, Chen L, et al. miR-671-5p inhibits epithelial-to-mesenchymal transition by downregulating FOXM1 expression in breast cancer. Oncotarget 2016;7:293-307. [Crossref] [PubMed]
- Gu Y, Wang W, Wang X, et al. Integrated network analysis identifies hsa-miR-4756-3p as a regulator of FOXM1 in Triple Negative Breast Cancer. Sci Rep 2019;9:13830. [Crossref] [PubMed]
- Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell 2007;128:763-76. [Crossref] [PubMed]
- Wei Y, Sun Q, Zhao L, et al. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med Oncol 2016;33:88. [Crossref] [PubMed]
- Luo X, Wang GH, Bian ZL, et al. Long non-coding RNA CCAL/miR-149/FOXM1 axis promotes metastasis in gastric cancer. Cell Death Dis 2018;9:993. [Crossref] [PubMed]
- Lv L, An X, Li H, et al. Effect of miR-155 knockdown on the reversal of doxorubicin resistance in human lung cancer A549/dox cells. Oncol Lett 2016;11:1161-6. [Crossref] [PubMed]
- Yu Q, Xu XP, Yin XM, et al. miR-155-5p increases the sensitivity of liver cancer cells to adriamycin by regulating ATG5-mediated autophagy. Neoplasma 2021;68:87-95. [Crossref] [PubMed]
- Zhang L, Chen T, Yan L, et al. MiR-155-3p acts as a tumor suppressor and reverses paclitaxel resistance via negative regulation of MYD88 in human breast cancer. Gene 2019;700:85-95. [Crossref] [PubMed]
- Yang LW, Wu XJ, Liang Y, et al. miR-155 increases stemness and decitabine resistance in triple-negative breast cancer cells by inhibiting TSPAN5. Mol Carcinog 2020;59:447-61. [Crossref] [PubMed]
- Eissa S, Matboli M, Shehata HH. Breast tissue-based microRNA panel highlights microRNA-23a and selected target genes as putative biomarkers for breast cancer. Transl Res 2015;165:417-27. [Crossref] [PubMed]
- Bayraktar R, Ivan C, Bayraktar E, et al. Dual Suppressive Effect of miR-34a on the FOXM1/eEF2-Kinase Axis Regulates Triple-Negative Breast Cancer Growth and Invasion. Clin Cancer Res 2018;24:4225-41. [Crossref] [PubMed]
- Tan X, Li Z, Ren S, et al. Dynamically decreased miR-671-5p expression is associated with oncogenic transformation and radiochemoresistance in breast cancer. Breast Cancer Res 2019;21:89. [Crossref] [PubMed]
- Li J, Meng H, Bai Y, et al. Regulation of lncRNA and Its Role in Cancer Metastasis. Oncol Res 2016;23:205-17. [Crossref] [PubMed]
- Renganathan A, Felley-Bosco E. Long Noncoding RNAs in Cancer and Therapeutic Potential. Adv Exp Med Biol 2017;1008:199-222. [Crossref] [PubMed]
- Ghafouri-Fard S, Taheri M. Long non-coding RNA signature in gastric cancer. Exp Mol Pathol 2020;113:104365 [Crossref] [PubMed]
- Milevskiy MJ, Al-Ejeh F, Saunus JM, et al. Long-range regulators of the lncRNA HOTAIR enhance its prognostic potential in breast cancer. Hum Mol Genet 2016;25:3269-83. [Crossref] [PubMed]
- Dong G, Pan T, Zhou D, et al. FBXL19-AS1 promotes cell proliferation and inhibits cell apoptosis via miR-876-5p/FOXM1 axis in breast cancer. Acta Biochim Biophys Sin (Shanghai) 2019;51:1106-13. [Crossref] [PubMed]
- Abba MC, Canzoneri R, Gurruchaga A, et al. LINC00885 a Novel Oncogenic Long Non-Coding RNA Associated with Early Stage Breast Cancer Progression. Int J Mol Sci 2020;21:7407. [Crossref] [PubMed]
- Hou Y, Zhu Q, Li Z, et al. The FOXM1-ABCC5 axis contributes to paclitaxel resistance in nasopharyngeal carcinoma cells. Cell Death Dis 2017;8:e2659 [Crossref] [PubMed]
- Yuan R, Hou Y, Sun W, et al. Natural products to prevent drug resistance in cancer chemotherapy: a review. Ann N Y Acad Sci 2017;1401:19-27. [Crossref] [PubMed]
- Ji X, Lu Y, Tian H, et al. Chemoresistance mechanisms of breast cancer and their countermeasures. Biomed Pharmacother 2019;114:108800 [Crossref] [PubMed]
- Li YJ, Lei YH, Yao N, et al. Autophagy and multidrug resistance in cancer. Chin J Cancer 2017;36:52. [Crossref] [PubMed]
- Kwok JM, Peck B, Monteiro LJ, et al. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mol Cancer Res 2010;8:24-34. [Crossref] [PubMed]
- Millour J, de Olano N, Horimoto Y, et al. ATM and p53 regulate FOXM1 expression via E2F in breast cancer epirubicin treatment and resistance. Mol Cancer Ther 2011;10:1046-58. [Crossref] [PubMed]
- Chen B, Lai J, Dai D, et al. PARPBP is a prognostic marker and confers anthracycline resistance to breast cancer. Ther Adv Med Oncol 2020;12:1758835920974212 [Crossref] [PubMed]
- Xin L, Zhou Q, Yuan YW, et al. METase/lncRNA HULC/FoxM1 reduced cisplatin resistance in gastric cancer by suppressing autophagy. J Cancer Res Clin Oncol 2019;145:2507-17. [Crossref] [PubMed]
- Li X, Qiu W, Liu B, et al. Forkhead box transcription factor 1 expression in gastric cancer: FOXM1 is a poor prognostic factor and mediates resistance to docetaxel. J Transl Med 2013;11:204. [Crossref] [PubMed]
- Lin JZ, Wang WW, Hu TT, et al. FOXM1 contributes to docetaxel resistance in castration-resistant prostate cancer by inducing AMPK/mTOR-mediated autophagy. Cancer Lett 2020;469:481-9. [Crossref] [PubMed]
- Varghese V, Magnani L, Harada-Shoji N, et al. FOXM1 modulates 5-FU resistance in colorectal cancer through regulating TYMS expression. Sci Rep 2019;9:1505. [Crossref] [PubMed]
- Ros S, Wright AJ, D'Santos P, et al. Metabolic Imaging Detects Resistance to PI3Kα Inhibition Mediated by Persistent FOXM1 Expression in ER+ Breast Cancer. Cancer Cell 2020;38:516-533.e9. [Crossref] [PubMed]
- Xie T, Geng J, Wang Y, et al. FOXM1 evokes 5-fluorouracil resistance in colorectal cancer depending on ABCC10. Oncotarget 2017;8:8574-89. [Crossref] [PubMed]
- Karunarathna U, Kongsema M, Zona S, et al. OTUB1 inhibits the ubiquitination and degradation of FOXM1 in breast cancer and epirubicin resistance. Oncogene 2016;35:1433-44. [Crossref] [PubMed]
- Nestal de Moraes G, Delbue D, Silva KL, et al. FOXM1 targets XIAP and Survivin to modulate breast cancer survival and chemoresistance. Cell Signal 2015;27:2496-505. [Crossref] [PubMed]
- Arceci A, Bonacci T, Wang X, et al. FOXM1 Deubiquitination by USP21 Regulates Cell Cycle Progression and Paclitaxel Sensitivity in Basal-like Breast Cancer. Cell Rep 2019;26:3076-3086.e6. [Crossref] [PubMed]
- Monteiro LJ, Khongkow P, Kongsema M, et al. The Forkhead Box M1 protein regulates BRIP1 expression and DNA damage repair in epirubicin treatment. Oncogene 2013;32:4634-45. [Crossref] [PubMed]
- Park YY, Jung SY, Jennings NB, et al. FOXM1 mediates Dox resistance in breast cancer by enhancing DNA repair. Carcinogenesis 2012;33:1843-53. [Crossref] [PubMed]
- Khongkow P, Karunarathna U, Khongkow M, et al. FOXM1 targets NBS1 to regulate DNA damage-induced senescence and epirubicin resistance. Oncogene 2014;33:4144-55. [Crossref] [PubMed]
- Khongkow P, Gomes AR, Gong C, et al. Paclitaxel targets FOXM1 to regulate KIF20A in mitotic catastrophe and breast cancer paclitaxel resistance. Oncogene 2016;35:990-1002. [Crossref] [PubMed]
- Hamurcu Z, Delibaşı N, Nalbantoglu U, et al. FOXM1 plays a role in autophagy by transcriptionally regulating Beclin-1 and LC3 genes in human triple-negative breast cancer cells. J Mol Med (Berl) 2019;97:491-508. [Crossref] [PubMed]
- Yang N, Wang C, Wang Z, et al. FOXM1 recruits nuclear Aurora kinase A to participate in a positive feedback loop essential for the self-renewal of breast cancer stem cells. Oncogene 2017;36:3428-40. [Crossref] [PubMed]
- Bergamaschi A, Madak-Erdogan Z, Kim YJ, et al. The forkhead transcription factor FOXM1 promotes endocrine resistance and invasiveness in estrogen receptor-positive breast cancer by expansion of stem-like cancer cells. Breast Cancer Res 2014;16:436. [Crossref] [PubMed]
- Teh MT. Cells brainwashed by FOXM1: do they have potential as biomarkers of cancer? Biomark Med 2012;6:499-501. [Crossref] [PubMed]
- Dai J, Yang L, Wang J, et al. Prognostic Value of FOXM1 in Patients with Malignant Solid Tumor: A Meta-Analysis and System Review. Dis Markers 2015;2015:352478 [Crossref] [PubMed]
- Bektas N. Tight correlation between expression of the Forkhead transcription factor FOXM1 and HER2 in human breast cancer. BMC Cancer 2008;8:42. [Crossref] [PubMed]
- Siraj AK, Pratheeshkumar P, Parvathareddy SK, et al. FoxM1 is an independent poor prognostic marker and therapeutic target for advanced Middle Eastern breast cancer. Oncotarget 2018;9:17466-82. [Crossref] [PubMed]
- Ahn H, Sim J, Abdul R, et al. Increased expression of forkhead box M1 is associated with aggressive phenotype and poor prognosis in estrogen receptor-positive breast cancer. J Korean Med Sci 2015;30:390-7. [Crossref] [PubMed]
- Abdeljaoued S, Bettaieb L, Nasri M, et al. Forkhead box M1 (FOXM1) expression predicts disease free survival and may mediate resistance to chemotherapy and hormonotherapy in male breast cancer. Breast Dis 2018;37:109-14. [Crossref] [PubMed]
- Alvarez-Fernández M, Medema RH. Novel functions of FoxM1: from molecular mechanisms to cancer therapy. Front Oncol 2013;3:30. [Crossref] [PubMed]
- Tan Y, Wang Q, Xie Y, et al. Identification of FOXM1 as a specific marker for triple-negative breast cancer. Int J Oncol 2019;54:87-97. [PubMed]
- Rajamanickam S, Panneerdoss S, Gorthi A, et al. Inhibition of FoxM1-Mediated DNA Repair by Imipramine Blue Suppresses Breast Cancer Growth and Metastasis. Clin Cancer Res 2016;22:3524-36. [Crossref] [PubMed]
- Kongsema M, Wongkhieo S, Khongkow M, et al. Molecular mechanism of Forkhead box M1 inhibition by thiostrepton in breast cancer cells. Oncol Rep 2019;42:953-62. [Crossref] [PubMed]
- Arora R, Yates C, Gary BD, et al. Panepoxydone targets NF-kB and FOXM1 to inhibit proliferation, induce apoptosis and reverse epithelial to mesenchymal transition in breast cancer. PLoS One 2014;9:e98370 [Crossref] [PubMed]
- Zhang N, Pati D. Separase Inhibitor Sepin-1 Inhibits Foxm1 Expression and Breast Cancer Cell Growth. J Cancer Sci Ther 2018;10:517. [Crossref] [PubMed]
- Wang K, Zhu X, Yin Y. Maslinic Acid Enhances Docetaxel Response in Human Docetaxel-Resistant Triple Negative Breast Carcinoma MDA-MB-231 Cells via Regulating MELK-FoxM1-ABCB1 Signaling Cascade. Front Pharmacol 2020;11:835. [Crossref] [PubMed]
- Ahmad A, Ali S, Wang Z, et al. 3,3'-Diindolylmethane enhances taxotere-induced growth inhibition of breast cancer cells through downregulation of FoxM1. Int J Cancer 2011;129:1781-91. [Crossref] [PubMed]
- Ahmad A, Ali S, Ahmed A, et al. 3, 3'-Diindolylmethane enhances the effectiveness of herceptin against HER-2/neu-expressing breast cancer cells. PLoS One 2013;8:e54657 [Crossref] [PubMed]
- Liu LP, Cao XC, Liu F, et al. Casticin induces breast cancer cell apoptosis by inhibiting the expression of forkhead box protein M1. Oncol Lett 2014;7:1711-7. [Crossref] [PubMed]
- Wang JS, Ren TN, Xi T. Ursolic acid induces apoptosis by suppressing the expression of FoxM1 in MCF-7 human breast cancer cells. Med Oncol 2012;29:10-5. [Crossref] [PubMed]
- Francis RE, Myatt SS, Krol J, et al. FoxM1 is a downstream target and marker of HER2 overexpression in breast cancer. Int J Oncol 2009;35:57-68. [PubMed]
- Gormally MV, Dexheimer TS, Marsico G, et al. Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition. Nat Commun 2014;5:5165. [Crossref] [PubMed]
- Ulhaka K, Kanokwiroon K, Khongkow M, et al. The Anticancer Effects of FDI-6, a FOXM1 Inhibitor, on Triple Negative Breast Cancer. Int J Mol Sci 2021;22:6685. [Crossref] [PubMed]
- Tabatabaei Dakhili SA, Pérez DJ, Gopal K, et al. SP1-independent inhibition of FOXM1 by modified thiazolidinediones. Eur J Med Chem 2021;209:112902 [Crossref] [PubMed]
- Pandit B, Gartel AL. FoxM1 knockdown sensitizes human cancer cells to proteasome inhibitor-induced apoptosis but not to autophagy. Cell Cycle 2011;10:3269-73. [Crossref] [PubMed]
- Halasi M, Hitchinson B, Shah BN, et al. Honokiol is a FOXM1 antagonist. Cell Death Dis 2018;9:84. [Crossref] [PubMed]
- Maharjan S, Kwon YS, Lee MG, et al. Cell cycle arrest-mediated cell death by morin in MDA-MB-231 triple-negative breast cancer cells. Pharmacol Rep 2021;73:1315-27. [Crossref] [PubMed]
- Ye X, Zhang Y, He B, et al. Quantitative proteomic analysis identifies new effectors of FOXM1 involved in breast cancer cell migration. Int J Clin Exp Pathol 2015;8:15836-44. [PubMed]
- Ziegler Y, Laws MJ, Sanabria Guillen V, et al. Suppression of FOXM1 activities and breast cancer growth in vitro and in vivo by a new class of compounds. NPJ Breast Cancer 2019;5:45. [Crossref] [PubMed]
- Dey P, Wang A, Ziegler Y, et al. Suppression of Tumor Growth, Metastasis, and Signaling Pathways by Reducing FOXM1 Activity in Triple Negative Breast Cancer. Cancers (Basel) 2020;12:2677. [Crossref] [PubMed]
- Ghandhariyoun N, Jaafari MR, Nikoofal-Sahlabadi S, et al. Reducing Doxorubicin resistance in breast cancer by liposomal FOXM1 aptamer: In vitro and in vivo. Life Sci 2020;262:118520 [Crossref] [PubMed]
- Wongkhieo S, Numdee K, Lam EWF, et al. Liposomal Thiostrepton Formulation and Its Effect on Breast Cancer Growth Inhibition. J Pharm Sci 2021;110:2508-16. [Crossref] [PubMed]
- Roßwag S, Cotarelo CL, Pantel K, et al. Functional Characterization of Circulating Tumor Cells (CTCs) from Metastatic ER+/HER2-Breast Cancer Reveals Dependence on HER2 and FOXM1 for Endocrine Therapy Resistance and Tumor Cell Survival: Implications for Treatment of ER+/HER2-Breast Cancer. Cancers (Basel) 2021;13:1810. [Crossref] [PubMed]
- O'Regan RM, Nahta R. Targeting forkhead box M1 transcription factor in breast cancer. Biochem Pharmacol 2018;154:407-13. [Crossref] [PubMed]
- Joshi T, Elias D, Stenvang J, et al. Integrative analysis of miRNA and gene expression reveals regulatory networks in tamoxifen-resistant breast cancer. Oncotarget 2016;7:57239-53. [Crossref] [PubMed]
- Hassani B, Taheri M, Asgari Y, et al. Expression Analysis of Long Non-Coding RNAs Related With FOXM1, GATA3, FOXA1 and ESR1 in Breast Tissues. Front Oncol 2021;11:671418 [Crossref] [PubMed]
- Sugie T. Immunotherapy for metastatic breast cancer. Chin Clin Oncol 2018;7:28. [Crossref] [PubMed]
- Keenan TE, Tolaney SM. Role of Immunotherapy in Triple-Negative Breast Cancer. J Natl Compr Canc Netw 2020;18:479-89. [Crossref] [PubMed]
- Franzoi MA, Romano E, Piccart M. Immunotherapy for early breast cancer: too soon, too superficial, or just right? Ann Oncol 2021;32:323-36. [Crossref] [PubMed]
- Yokomine K, Senju S, Nakatsura T, et al. The forkhead box M1 transcription factor as a candidate of target for anti-cancer immunotherapy. Int J Cancer 2010;126:2153-63. [Crossref] [PubMed]
- Zhang S, Liu S, Liu X, et al. Identification of JAK2 and FOXM1 expression as novel candidate biomarkers for predicting the benefit of immunotherapy in lung squamous cell carcinoma. Ann Transl Med 2021;9:1081. [Crossref] [PubMed]
- Su H, Li B, Zheng L, et al. Immunotherapy based on dendritic cells pulsed with CTPFoxM1 fusion protein protects against the development of hepatocellular carcinoma. Oncotarget. 2016;7:48401-11. [Crossref] [PubMed]
- Di Sante G, Page J, Jiao X, et al. Recent advances with cyclin-dependent kinase inhibitors: therapeutic agents for breast cancer and their role in immuno-oncology. Expert Rev Anticancer Ther 2019;19:569-87. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


