
This article has an erratum available at: http://dx.doi.org/10.21037/atm-2024-6 the article has been update on 2024-05-23 at here.
LncRNA NFYC-AS1 promotes the development of lung adenocarcinomas through autophagy, apoptosis, and MET/c-Myc oncogenic proteins
Introduction
Lung cancer is one of the most commonly diagnosed cancers and the leading cause of cancer death globally (1). The overall survival (OS) of advanced lung cancer was only 10–12 months in the absence of targeted drugs about 10 years ago. With the continuous application of more and more targeted drugs, the OS of advanced lung cancer patients treated with targeted therapy was extended to more than 3 years (2,3). The same trend was seen in early stage lung cancer patients with EGFR mutations who took EGFR TKIs as adjuvant chemotherapy (4,5). The premise of targeted therapy relies on the discovery of more targeted driver genes.
Long noncoding RNAs (lncRNAs) are a class of RNA molecules. They are not translated into proteins and are typically over 200 nucleotides long. LncRNAs are intimately involved in the development of cancer (6-9). Aberrantly expressed lncRNAs have been found to be oncogenes of various types of cancers (10-12). However, the functions of most lncRNAs remain unclear. Clarification of their functions and mechanisms in carcinogenesis can potentially offer new therapeutic targets (13).
Li et al. analyzed the RNA-seq data and miRNA-seq data of lung adenocarcinoma (LAUD) from The Cancer Genome Atlas (TCGA) database to identify critical lncRNAs and determine the molecular pathogenesis. Nuclear transcription factor Y subunit C antisense RNA 1 (NFYC-AS1) was revealed to be a potential prognostic biomarker (14). However, the authors did not further verify the function of NFYC-AS1 in lung cancer cell lines. There have been few studies regarding the function of NFYC-AS1. For instance, van der Plaat et al. found that NFYC-AS1 may play a role in airflow obstruction in never smokers by analyzing the genome-wide association study (GWAS) data (15). However, the authors also did not further verify the function of NFYC-AS1 in cell lines or animal models. So far, there has been no data available on the molecular function, phenotypes, animal models, miRNA, transcription factor targets, or HOMER transcription, among others, of the NFYC-AS1 gene. The following molecular assays demonstrated that NFYC-AS1 may promote the proliferation of LAUD through autophagy and apoptosis, and MET/c-Myc oncogenic proteins.
It was reported showed that autophagy in cancer serves as both a tumor suppressor and promoter (16). Targeting autophagy-related pathways may be a promising strategy for cancer therapy. As we all know, apoptosis plays a critical role in tumorigenesis. MET has been found to be often deregulated in solid malignancies, which in turn promotes tumor growth, progression and invasion (17). c-Myc is a human oncogene, which is frequently deregulated in human cancers and widely involved in tumorigenesis (18). We present the following article in accordance with the MDAR checklist (available at https://dx.doi.org/10.21037/atm-21-4995).
Methods
Cell culture
LAUD cell lines (H1299/PC9/A549/H838) and normal LAUD cell line (HBE) were purchased from Sigma Chemical (ATCC, China). All media were supplemented with 10% fetal bovine serum (FBS, Invitrogen, Guangzhou, China), unless otherwise stated.
Cell proliferation assays
Cells were seeded onto 96-well plates at a density of 1,000 cells per well. At the 24- and 48-hour time points, small interfering RNA (siRNA, of NFYC-AS1 and control) and transfection reagents were added to the cells. At 96–120 hours after transfection, cell proliferation was measured with the WST-1 reagent (Roche, Mannheim, Germany) following the instructions from the manufacturer. All experiments were performed in triplicate.
Colony formation assay
The cells were inoculated onto 6-well plates at a density of 5×103 cells/well, and were subsequently cultured for 10 days. The cells were then fixed with formalin followed by staining with crystal violet. The numbers of colonies were then counted and the reported colony numbers were relative values.
RNA isolation and quantitative reverse transcription PCR (qRT-PCR)
Total RNA was isolated from cells (tissue or cultured) using the miRNeasy Mini kit (Qiagen) following the instructions from the manufacturer. The reverse transcription reaction was performed with the High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific) using 1 µg of total RNA in a final volume of 20 µL under standard conditions and using random primers. Subsequently, the qRT-PCR reaction was carried out using 1 µL of the corresponding cDNA with Power SYBR Green master Mix (Thermo Fisher Scientific) following the instructions from the manufacturer. The results were normalized based on the expression of GADPH. The PCR protocol was 15 sec of 95 °C + 60 sec of 60 °C repeated for 40 cycles, performed on the StepOne Real-Time PCR System (Thermo Fisher Scientific). The following primer sequences are used: NFYC-AS1: forward: 5'-AGTGAGGTCTCTGGCGAAGGC-3'; reverse: 5'-CCTGCTCTTCCTGGACTCCTGAG-3'. All procedures were performed in triplicate and data were analyzed using the comparative cycle threshold (CT) method.
Cell migration and invasion assay
Quantitative analysis of the migration and invasion capabilities of LAUD cells was performed using the transwell chamber system. A total of 60 µL of diluted extracellular matrix (ECM) gel solution was added to the upper chambers (Costar Inc., USA) and then incubated for 4 hours at 37 °C. For the migration assay, the upper chambers were prepared using the same protocol, except that ECM was not added. Subsequently, cells were seeded at a density of 1×105 cells in 100 µL medium with 1% FBS in each well. The lower chambers were filled with 500 µL RPMI and DMEM containing 10% FBS. The transwells were then incubated for 24–48 hours at 37 °C with 5% CO2 to allow cell migration. When the incubation period ended, the remaining cells in the upper chamber were removed with a cotton swab. Cells that reached the bottom of the membrane were then fixed with paraformaldehyde. The fixed cells were stained by adding crystal violet and incubated at room temperature for 30 min. The crystal violet bound on the cells was washed off with 200 µL of 33% glacial acetic acid, and the absorbance of the eluent at 590 nm was measured to determine the cell number.
Flow cytometric assay
Cells were plated into 6-well plates at 6×104 cells/well. After culturing the siRNA-transfected cells for 48 hours, flow cytometry analysis was performed. Fluorescein isothiocyanate-Annexin V and propidium iodide (Absin, Shanghai, China) were used to stain 300 µL binding buffer of cell suspension. Apogee Flow Cytometers (Apogee Flow Systems, Hemel Hempstead, UK) were used to analyze the stained cells.
Western blotting
At 72 hours after siRNA transfection, cells were harvested and subjected to lysis, electrophoresis, and target protein visualization following a previously described protocol (19). Total cell lysates were prepared in sample buffer and boiled for 5 min at 95 °C. The sample was then subjected to SDS-PAGE for 3 hours at 80 V, and transferred to PVDF membranes for another 3 hours. The membranes were then incubated at 4 °C overnight with specific antibodies for p62, Beclin 1, MET, c-Myc, and GAPDH. Subsequently, the membranes were washed 3 times with 1% TBST, incubated for 1 hour with secondary antibodies, developed using ECL, and exposed to X-ray film.
Statistical analysis
Data analysis was performed using SPSS. All variables were assumed to follow a continuous normal distribution. Data such as proliferation were evaluated by an unpaired Student’s t-test. All statistical values were expressed as mean ± SD. All statistical tests were performed at a level of significance of α=0.05. Unless stated otherwise, all cell biological experiments were independently triplicated.
Results
NFYC-AS1 promotes the proliferation and colony formation of LAUD cells
Up to now, there has been no report on the NFYC-AS1 expression status in cancers. We performed qRT-PCR to determine NFYC-AS1 expression in 4 LAUD cell lines (H1299/PC9/A549/H838) and 1 normal lung cell line (HBE) as the control. We confirmed that the expression of NFYC-AS1 was higher in LAUD cell lines (Figure 1A). We chose the H1299 and PC9 cell lines, which had the highest expression of NFYC-AS1, to conduct the follow-up experiments.
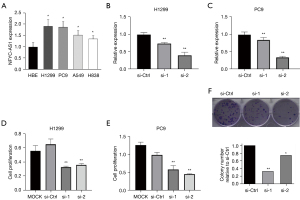
To explore the function of NFYC-AS1, we firstly performed siRNA-mediated knockdown of NFYC-AS1 in H1299 and PC9 cells. The expression of NFYC-AS1 was significantly reduced after transfection with NFYC-AS1 siRNAs as determined by qRT-PCR assays (Figure 1B,1C).
We performed the WST-1 assay to investigate the impact of NFYC-AS1 on cell proliferation. The proliferation of H1299 and PC9 cells was inhibited by 35–50% after NFYC-AS1 knockdown (Figure 1D,1E). We also performed colony formation assays in H1299 and PC9 cells. The PC9 cell line was unable to form colonies, while the colony formation of H1299 cells was significantly inhibited. The clone formation ability of si-1 and si-2 was inhibited by 26–68% after the knockdown of NFYC-AS1 in H1299 cell line (Figure 1F). The results suggested that NFYC-AS1 could promote the proliferation and colony formation of LAUD cells.
NFYC-AS1 promotes the invasion and migration of LAUD cells
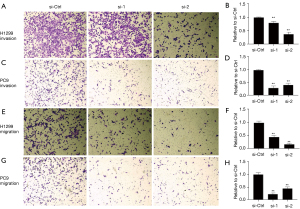
We performed transwell assays to explore whether NFYC-AS1 affects the migration and invasion of H1299 and PC9 cells. We found that cell invasion was inhibited by 21–70% after NFYC-AS1 knockdown (Figure 2A-2D), and migration was decreased by 55–83% after NFYC-AS1 knockdown (Figure 2E-2H). The results suggested that NFYC-AS1 could promote the migration and invasion of LAUD cells.
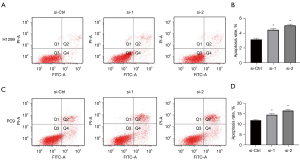
NFYC-AS1 inhibits the apoptosis of LAUD cells
To probe the potential role of NFYC-AS1 in cell apoptosis regulation, apoptosis was analyzed by flow cytometry analysis. The results showed that cells transfected with NFYC-AS1 siRNA had a higher rate of apoptosis compared with that in control cells (Figure 3). The results suggested that NFYC-AS1 could inhibit the apoptosis of LAUD cells.
The downstream proteins of NFYC-AS1
We detected the expression of certain proteins in the H1299 and PC9 cell lines to explore the downstream signaling proteins of NFYC-AS1. In terms of the expression of major autophagy proteins, we found that p62 was downregulated while Beclin 1 was upregulated in both cell lines after NFYC-AS1 knockdown. However, LC3B expression was not changed (Figure 4A,4B). This illustrates that NFYC-AS1 may function through autophagy, but not through the LC3B protein. The proteins p53 and PARP were upregulated after NFYC-AS1 knockdown (Figure 4C,4D). The cleaved PARP bands were present in the western blot of H1299 cells, but the same bands were not observed for PC9 cells. This suggested that NFYC-AS1 might promote apoptosis, which was consistent with the results of flow cytometry analysis.
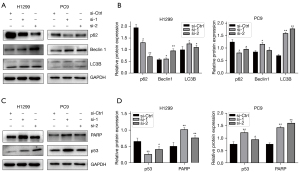
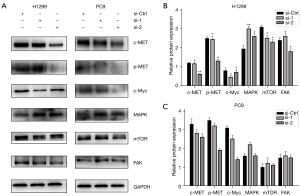
The expression levels of the oncogenic proteins MET and c-Myc were downregulated after NFYC-AS1 knockdown (Figure 5). This illustrates that MET and c-Myc may play important roles in LAUD cells through the NFYC-AS1 network. The p-mTOR and FAK were not changed after NFYC-AS1 knockdown, which suggests that NFYC-AS1 does not function via FAK and p-mTOR. The MAPK/ERK was upregulated (Figure 5), which we could not explain.
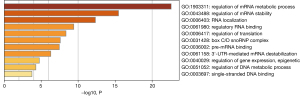
We performed DAVID gene ontology/pathway analysis of NFYC-AS1 correlated (Pearson correlation) genes based on TCGA data and observed two pathways that were involved in LAUD, namely the regulation of mRNA metabolic process and the regulation of mRNA stability (Figure 6).
Discussion
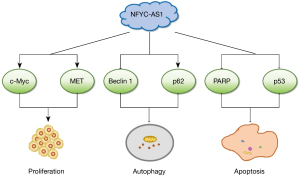
Antisense RNAs are generally thought to have a regulatory role, however, the function of NFYC-AS1 is still unknown. Our results showed that the proliferation, migration, and invasion of H1299 and PC9 cells were inhibited while apoptosis was increased after NFYC-AS1 knockdown. Apoptosis and autophagy were activated. Furthermore, the oncogenic protein expression levels of MET and c-Myc were downregulated. We have summarized the aforementioned results in the schematic diagram in Figure 7.
Nuclear factor-Y (NF-Y), also termed CMP-binding factor (CBF) or cysteine proteinase-1 (CP1), is a mammalian transcription factor critical in regulating the expression of many genes via binding to the CCAAT box. The members of the NF-Y family are trimers each constituting three subunits NF-YA, NF-YB, and NF-YC (20). NFYC-AS1 is a lncRNA that is the antisense transcript of the NF-YC subunit. Antisense lncRNAs have been recognized to play a regulatory role in the expression of corresponding coding genes at the post-transcriptional level (21) and, by regulating both oncogenes and anti-oncogenes, actively participate in carcinogenesis. To date, however, no molecular biological study has been conducted to explore the function of NFYC-AS1. We found that the proliferation, migration, and invasion of H1299 and PC9 cells were inhibited and apoptosis was increased after NFYC-AS1 knockdown, which indicated that NFYC-AS1 might be involved in the regulation of the proliferation and metastasis of LAUD. Many other lncRNAs, such as HOTTIP (22,23) and HOTAIR (24-26), have been proven to be oncogenes in cancers. To our knowledge, this is the first study proving that NFYC-AS1 has the characteristics of a driver gene.
Then, we detected the proteins that may participate in the regulation of the functions verified above. In terms of the major autophagy proteins, p62 was downregulated and Beclin 1 was upregulated, while LC3B was not changed after NFYC-AS1 knockdown. Autophagy is a sophisticated cellular process under the regulation of over 30 autophagy-related genes, among which Beclin 1, LC3B, and p62 play significant roles. Samples that test positive for 2+ of these 3 proteins are regarded as having autophagy activation (27,28). Our results illustrated that NFYC-AS1 might activate autophagy, but not through the LC3B protein. The role of autophagy in cancer is considered to be a double-edged sword. Under different circumstances, autophagy may play a role in promoting either survival or apoptosis (29,30). Our results suggest that NFYC-AS1 may promote the proliferation of LAUD cells by inhibiting autophagy.
Apoptosis, which is genetically programmed cell death, plays a critical role both in normal development and in tumorigenesis. LAUD cells had a higher rate of apoptosis after NFYC-AS1 knockdown. The apoptosis-related proteins p53 and PARP were upregulated after NFYC-AS1 knockdown. p53 is a gene that, in its unmutated form, regulates the transcription of numerous genes that play roles in various cellular processes to suppress tumorigenesis. Its mutation is associated with human cancer more than any other gene (31). In recent years, the important role played by p53 in initiating apoptosis under various stress stimuli has been increasingly brought to light (32-34). PARP is the key modulator of apoptosis, through the cleavage of PARP (35,36). In summary, NFYC-AS1 may inhibit apoptosis via p53 and PARP.
MET and c-Myc are primary oncogenic drivers. Aberrant MET and c-Myc expression drives tumor growth through increased cell proliferation, survival, invasion, and metastasis (37-39). The knockdown of NFYC-AS1 was related to the downregulation of MET and c-Myc, which suggests that the promotive role of NFYC-AS1 in tumorigenesis is associated with MET and c-Myc. FAK is a protein tyrosine kinase that regulates cellular adhesion, motility, proliferation, and survival in various types of cells. mTOR plays a central role in regulating cell growth, proliferation, and survival. Our results showed that the expression of FAK and p-mTOR were not changed after NFYC-AS1 knockdown, which indicates that NFYC-AS1 does not function via FAK and p-mTOR. MAPK/ERK has been proven to contribute to cancer progression. Our results showed that the expression of MAPK was upregulated after NFYC-AS1 knockdown, which we could not explain.
In summary, NFYC-AS1 promotes the proliferation, migration, and invasion and suppresses the apoptosis of LAUD cells. The possible mechanisms may include the induction of autophagy and the downregulation of oncoproteins such as MET and c-Myc. These findings suggest that NFYC-AS1 is a functional lncRNA in human LAUD cells. NFYC-AS1 may promote the proliferation of LAUD through autophagy and apoptosis.
Acknowledgments
Funding: This work was supported in part by National Natural Science Foundation of China (NSFC) (Grant No. 81702270 to FN, 81802335 to YS), Affiliated Hospital of Guangzhou Medical University Foundation (Grant No. 2013A41 to JD), and Clinical Key Specialty Construction Project of Guangzhou Medical University (Grant No. YYPT202017).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://dx.doi.org/10.21037/atm-21-4995
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-4995
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-4995). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998-2006. [Crossref] [PubMed]
- Liu Y, Xiong ZC, Sun X, et al. Impact of apatinib in combination with osimertinib on EGFR T790M-positive lung adenocarcinoma. Transl Cancer Res 2019;8:2151-63. [Crossref]
- Zhong WZ, Chen KN, Chen C, et al. Erlotinib Versus Gemcitabine Plus Cisplatin as Neoadjuvant Treatment of Stage IIIA-N2 EGFR-Mutant Non-Small-Cell Lung Cancer (EMERGING-CTONG 1103): A Randomized Phase II Study. J Clin Oncol 2019;37:2235-45. [Crossref] [PubMed]
- Xu ST, Xi JJ, Zhong WZ, et al. The Unique Spatial-Temporal Treatment Failure Patterns of Adjuvant Gefitinib Therapy: A Post Hoc Analysis of the ADJUVANT Trial (CTONG 1104). J Thorac Oncol 2019;14:503-12. [Crossref] [PubMed]
- Kulkarni S, Lied A, Kulkarni V, et al. CCR5AS lncRNA variation differentially regulates CCR5, influencing HIV disease outcome. Nat Immunol 2019;20:824-34. [Crossref] [PubMed]
- Hu Q, Ye Y, Chan LC, et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol 2019;20:835-51. [Crossref] [PubMed]
- Chen F, Chen J, Yang L, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol 2019;21:498-510. [Crossref] [PubMed]
- Wu J, Zhu P, Lu T, et al. The long non-coding RNA LncHDAC2 drives the self-renewal of liver cancer stem cells via activation of Hedgehog signaling. J Hepatol 2019;70:918-29. [Crossref] [PubMed]
- Yu M, Xue Y, Zheng J, et al. Linc00152 promotes malignant progression of glioma stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway. Mol Cancer 2017;16:110. [Crossref] [PubMed]
- Wu Z, Ouyang C, Peng L. Risk assessment model and nomogram established by differentially expressed lncRNAs for early-stage lung squamous cell carcinoma. Transl Cancer Res 2020;9:5304-14. [Crossref]
- Yang J, Qiu Q, Qian X, et al. Long noncoding RNA LCAT1 functions as a ceRNA to regulate RAC1 function by sponging miR-4715-5p in lung cancer. Mol Cancer 2019;18:171. [Crossref] [PubMed]
- Esposito R, Bosch N, Lanzós A, et al. Hacking the Cancer Genome: Profiling Therapeutically Actionable Long Non-coding RNAs Using CRISPR-Cas9 Screening. Cancer Cell 2019;35:545-57. [Crossref] [PubMed]
- Li DS, Ainiwaer JL, Sheyhiding I, et al. Identification of key long non-coding RNAs as competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma. Eur Rev Med Pharmacol Sci 2016;20:2285-95. [PubMed]
- van der Plaat DA, Vonk JM, Lahousse L, et al. Limited overlap in significant hits between genome-wide association studies on two airflow obstruction definitions in the same population. BMC Pulm Med 2019;19:58. [Crossref] [PubMed]
- Li L, Liu WL, Su L, et al. The Role of Autophagy in Cancer Radiotherapy. Curr Mol Pharmacol 2020;13:31-40. [Crossref] [PubMed]
- Gherardi E, Birchmeier W, Birchmeier C, et al. Targeting MET in cancer: rationale and progress. Nat Rev Cancer 2012;12:89-103. [Crossref] [PubMed]
- Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene 2001;20:5595-610. [Crossref] [PubMed]
- Wang L, He Y, Liu W, et al. Non-coding RNA LINC00857 is predictive of poor patient survival and promotes tumor progression via cell cycle regulation in lung cancer. Oncotarget 2016;7:11487-99. [Crossref] [PubMed]
- Tong Y, Merino D, Nimmervoll B, et al. Cross-Species Genomics Identifies TAF12, NFYC, and RAD54L as Choroid Plexus Carcinoma Oncogenes. Cancer Cell 2015;27:712-27. [Crossref] [PubMed]
- Katayama S, Tomaru Y, Kasukawa T, et al. Antisense transcription in the mammalian transcriptome. Science 2005;309:1564-6. [Crossref] [PubMed]
- Zhao R, Zhang Y, Zhang X, et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer 2018;17:68. [Crossref] [PubMed]
- Luo H, Zhu G, Xu J, et al. HOTTIP lncRNA Promotes Hematopoietic Stem Cell Self-Renewal Leading to AML-like Disease in Mice. Cancer Cell 2019;36:645-659.e8. [Crossref] [PubMed]
- Wasson CW, Abignano G, Hermes H, et al. Long non-coding RNA HOTAIR drives EZH2-dependent myofibroblast activation in systemic sclerosis through miRNA 34a-dependent activation of NOTCH. Ann Rheum Dis 2020;79:507-17. [Crossref] [PubMed]
- Yang L, Peng X, Li Y, et al. Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol Cancer 2019;18:78. [Crossref] [PubMed]
- Zhang L, He A, Chen B, et al. A HOTAIR regulatory element modulates glioma cell sensitivity to temozolomide through long-range regulation of multiple target genes. Genome Res 2020;30:155-63. [Crossref] [PubMed]
- Mathew R, Karp CM, Beaudoin B, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009;137:1062-75. [Crossref] [PubMed]
- Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 2009;137:1001-4. [Crossref] [PubMed]
- Guo JY, White E. Autophagy, Metabolism, and Cancer. Cold Spring Harb Symp Quant Biol 2016;81:73-8. [Crossref] [PubMed]
- Liu J, Fan L, Wang H, et al. Autophagy, a double-edged sword in anti-angiogenesis therapy. Med Oncol 2016;33:10. [Crossref] [PubMed]
- Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 2014;25:304-17. [Crossref] [PubMed]
- Bohlman S, Manfredi JJ. Mdm2-RNA Interactions as a Target for Cancer Therapy: It's Not All About p53. Cancer Cell 2016;30:513-4. [Crossref] [PubMed]
- Li Y, Cao Y, Xiao J, et al. Inhibitor of apoptosis-stimulating protein of p53 inhibits ferroptosis and alleviates intestinal ischemia/reperfusion-induced acute lung injury. Cell Death Differ 2020;27:2635-50. [Crossref] [PubMed]
- Saller E, Tom E, Brunori M, et al. Increased apoptosis induction by 121F mutant p53. EMBO J 1999;18:4424-37. [Crossref] [PubMed]
- Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917-21. [Crossref] [PubMed]
- Altmeyer A, Simmons RC, Krajewski S, et al. Reversal of EBV immortalization precedes apoptosis in IL-6-induced human B cell terminal differentiation. Immunity 1997;7:667-77. [Crossref] [PubMed]
- Cortot AB, Kherrouche Z, Descarpentries C, et al. Exon 14 Deleted MET Receptor as a New Biomarker and Target in Cancers. J Natl Cancer Inst 2017; [Crossref] [PubMed]
- Awad MM, Oxnard GR, Jackman DM, et al. MET Exon 14 Mutations in Non-Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J Clin Oncol 2016;34:721-30. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


