
Trimethylamine-N-oxide-stimulated hepatocyte-derived exosomes promote inflammation and endothelial dysfunction through nuclear factor-kappa B signaling
Introduction
Ischemic heart disease remains a major long-term public health challenge around the world (1). Vascular inflammation and endothelial dysfunction are characteristics of atherosclerosis (2). In recent years, trimethylamine-N-oxide (TMAO) has been proven to be a proatherogenic compound that exerts pathogenic effects by promoting vascular inflammation and endothelial dysfunction (3-7). However, the molecular mechanisms underlying TMAO have not yet been completely explained.
Exosomes (Exos) are nanosized membrane particles, 50 to 100 nm size in range, which are secreted by various types of cells and transmit information from cells to cells. The functions and characteristics of Exos mainly depend on the types and states of the host cells from which they originated. The proteins, lipids, micro ribonucleic acids (miRNAs), and other non-coding RNAs carried by Exos are thought to be the key components for intercellular communication, and thus play crucial roles in many biological processes, such as immune response, cardiovascular disease, tumor, and neurodegenerative disease (8-10).
Recent studies have shown that hepatocyte-derived Exos play an important role in the regulation of vascular inflammation and endothelial function (11-13). As TMAO is produced in the liver (14), hepatocytes may be the first potential target for TMAO. Actually, recent research has shown that TMAO may work directly on hepatocytes, and thus exert an influence over metabolic syndrome (15). However, it is not yet clear whether TMAO can directly stimulate hepatocytes to produce Exos to mediate the unfavorable effects of TMAO on vascular endothelial cells (VECs).
In this study, we found that TMAO at a physiological concentration can indirectly talk to VECs by promoting hepatocytes to produce Exos that enriched with differential miRNAs. Specifically, Exos derived from TMAO-stimulated hepatocytes, are found to promote the expressions of inflammatory markers and cell apoptosis, impair cell migration, and inhibit endothelium-dependent vasodilation. Although studies have shown that both TMAO and hepatocyte-derived Exos were involved with inflammation and endothelial function (3-5,11-13), our findings firstly uncover a direct relationship between the two and provide novel insights into the interaction between liver and vasculature in atherogenesis provoked by TMAO. We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-5043).
Methods
AML12 cell cultures, treatment, Exo isolation, and characterization
As the source of normal primary hepatocytes is very limited, alpha mouse liver 12 (AML12) cells are widely used in research, as they closely phenocopy the normal primary hepatocytes (15-17). Hence, we adopted the hepatocyte model in this study. AML12 cells (iCell Bioscience Inc., Shanghai) were cultured in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (iCell Bioscience Inc., Shanghai) containing Exo-depleted serum (ViVaCell, Shanghai), and treated with TMAO (Tokyo Chemical Industry Co., Ltd.) at a physiological concentration of 50 µmol/L. The untreated group served as the control group. After 48 h, the Exos were isolated and purified from the culture supernatant using differential centrifugation. Briefly, the medium was collected and centrifuged at 300 ×g for 10 min, 2,000 ×g for 10 min at 4 °C, and then again at 10,000 ×g for 30 min at 4 °C. The supernatant was passed through a 0.22 µm filter (Millipore) and ultracentrifuged at 110,000 ×g for 70 min at 4 °C. The pellets were then washed with phosphate-buffered saline (PBS) followed by another ultracentrifugation at 110,000 ×g for 70 min at 4 °C and then resuspended in PBS. The protein levels of the Exos were measured using a bicinchoninic acid (BCA) protein assay kit (23228, Thermo Scientific). The ultrastructure and size distribution of the Exos were identified by transmission electron microscopy (JEM1200-EX, Japan) and nanoparticle tracking analysis (Nanosight NS300, Malvern, UK), respectively. The protein markers of cluster of differentiation 81 (CD81) and tumor susceptibility gene 101 (TSG101) were detected by western blotting. The Exos were labelled with 1,1'-dioctadecyl-3,3,3'3'-tetramethylindocarbocyanine perchlorate (DiI) for the in vitro tracer experiment. Briefly, 250 µg Exos were labelled with 1 µM DiI (Beyotime Biotechnology), and incubated at 37 °C for 5 min, then Exos-depleted serum was added to counteract the excess dye. After washing with PBS, the labelled Exos were isolated by ultracentrifugation as described above. The labelled Exos were incubated with human aortic endothelial cells (HAECs) for 24 h, and the cells were then fixed and visualized with a confocal fluorescence microscope.
Endothelial cell culture and treatment
HAECs (iCell Bioscience Inc, Shanghai) were cultured in endothelial cell medium (ScienCell) supplemented with 5% fetal bovine serum, 1% growth factors, and 1% penicillin/streptomycin. The cells were treated with Exos isolated from TMAO-stimulated group (TMAO-Exos) and TMAO-free group (Control-Exos) at a concentration of 1:100 (v/v).
Western blot
The procedure was performed according to standard protocols as previously described (18). The protein concentration was determined using a BCA protein assay kit (23228, Thermo Scientific). The samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto Millipore polyvinylidene difluoride membranes. CD81 primary antibody was purchased from Servicebio (Wuhan, China), TSG101 was purchased from ZEN BIO (Chengdu, China), nuclear factor-kappa B (NF-κB) p65 and Phospho-NF-κB p65 (p-NF-κB p65) at serine536 (Ser536) were purchased from Cell Signaling Technology (Danvers, MA, USA), and glyceraldehyde phosphate dehydrogenase (GAPDH) was purchased from Proteintech Group (Rosemont, IL, USA). The proteins were visualized with enhanced chemiluminescence reagent (Millipore, MA, USA).
Quantitative polymerase chain reaction (qPCR)
qPCR was performed according to standard protocols as previously described (19). Briefly, total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific, USA), and the concentration was measured using NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). For messenger RNA (mRNA), RNA was reversely transcribed into complementary deoxyribonucleic acid (cDNA) using Color Reverse Transcription Kit (EZBioscience, USA), and qPCR was performed on Bio-Rad CFX-96 (Bio-Rad, USA) with Color SYBR Green qPCR Master Mix (EZBioscience, USA). The expressions of interleukin-6 (IL-6), monocyte chemotactic protein-1 (MCP-1), and tumor necrosis factor-α (TNF-α) were normalized to GAPDH. The qPCR primers used in the study are listed in Table 1. For miRNA, the RNA was reversely transcribed into cDNA using Reverse Transcriptase M-MLV (RNase H-) (Takara, Japan). qPCR was performed on Applied Biosystems ViiATM7 Real-Time PCR System (ABI, USA) with AceQ qPCR SYBR Green Master Mix (Vazyme Biotech, Nanjing, China). The expressions of miR-302d-3p, miR-302b-3p, miR-302a-3p, miR-103-3p, 6_9856 (prediction), miR-199b-3p, miR-199a-3p, miR-92a-3p, miR-122-5p, and miR-744-5p were normalized to snoU6. The qPCR primers used in the study are listed in Table 2.
Table 1
| Name | Forward sequence | Reverse sequence |
|---|---|---|
| IL-6 | TGGCAGAAAACAACCTGAACCTT | TCTGGCTTGTTCCTCACTACTCT |
| MCP-1 | CTCATAGCAGCCACCTTCATTCC | GATCACAGCTTCTTTGGGACACT |
| TNF-α | CTCAGCCTCTTCTCCTTCCTGAT | TCGAGAAGATGATCTGACTGCCT |
| GAPDH | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAG |
qPCR, quantitative polymerase chain reaction; IL-6, interleukin-6; MCP-1, monocyte chemotactic protein-1; TNF-α, tumor necrosis factor-α; GAPDH, glyceraldehyde phosphate dehydrogenase.
Table 2
| Name | Primer | Sequence (5’-3’) |
|---|---|---|
| miR-302d-3p | RT-primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACACTCAAA |
| Forward primer | ATGGTTCGTGGGGTGCTTCCATGTTTGAGTGT | |
| miR-302b-3p | RT-primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACTAAA |
| Forward primer | ATGGTTCGTGGGGTGCTTCCATGTTTTAGTAG | |
| miR-302a-3p | RT-primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCACCAAA |
| Forward primer | ATGGTTCGTGGGGTGCTTCCATGTTTTGGTGA | |
| miR-103-3p | RT-primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCATAGCC |
| Forward primer | ATGGTTCGTGGGAGCAGCATTGTACAGGG | |
| #6_9856 (prediction) | RT-primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAAAGCCA |
| Forward primer | ATGGTTCGTGGGGGTGTACTGGCTT | |
| miR-199b-3p | RT-primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTAACCAAT |
| Forward primer | ATGGTTCGTAAATTGTACAGTAGTCT | |
| miR-199a-3p | RT-primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTAACCAAT |
| Forward primer | ATGGTTCGTCCGTTGTACAGTAGTCT | |
| miR-92a-3p | RT-primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAGGCCGGGA |
| Forward primer | GCACTTGTCCCGGCCTG | |
| miR-122-5p | RT-primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAAACACC |
| Forward primer | ATGGTTCGTGGGTGGAGTGTGACAATGGT | |
| miR-744-5p | RT-primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGCTGTTA |
| Forward primer | GCGGGGCTAGGGCTAAC | |
| Universal primer (reverse primer) | GTGCAGGGTCCGAGGTATT | |
| snoU6 | Forward primer | CTCGCTTCGGCAGCACA |
| Reserves Primer | AACGCTTCACGAATTTGCGT | |
#, a newly predicted miRNA with function unknown. qPCR, quantitative polymerase chain reaction.
Cell apoptosis
Cell apoptosis was evaluated using Hoechst 33342 staining solution (Sigma) and flow cytometry assay, respectively. For Hoechst staining, the treated HAECs were washed twice with PBS and fixed with 4% paraformaldehyde for 10 min, and washed twice with PBS again. Hoechst 33342 staining solution (10 µg/mL) was then added to cover the cells, and after incubating for 8 min in the dark, the cells were washed twice with PBS. Next, the cells were observed under a fluorescent microscope (Leica DMI4000B, Germany). For flow cytometry assay, the treated HAECs were detached with 0.25% non-EDTA trypsin and washed with PBS and 1× binding buffer. Then cells with a density of 1×105 were resuspended in 100 µL 1× binding buffer and incubated with 5 µL Annexin V-FITC and 5 µL propidium iodide (PI) at room temperature away from light for 15 min according to the manufacturer’s manual. The cells were added 300 µL 1× binding buffer and the apoptosis ratios were detected by flow cytometry (CytoFLEX, Beckman Coulter, USA).
Cell scratch assay and transwell assay
For scratch assay, HAECs were seeded on 6-well plates and grown with Exos until they reached 90% confluence. The cell monolayer was scraped in a straight line to create a “scratch” with a sterile p200 pipette. Debris was removed gently by washing with PBS. Next, the cells continued to be incubated with the Exos, and images at 0 and 12 h were captured using an inversion fluorescence microscope. For transwell assay, the treated HAECs were seeded to Transwell chambers (Corning, NY, USA) with a density of 2×104, the chamber was placed in a 24-well plate and incubated for 12 h at 37 °C. Non-attached cells were gently removed with PBS, and adherent cells were fixed with 4% paraformaldehyde for 15 min and 0.3% crystal violet for another 15 min, and counted using inversion microscope.
Vasodilation study
The animal experiments were performed under a project license (No. S2020-168) granted by the ethics committees for animal experiments of Guangzhou Medical University, in compliance with the National Research Council’s Guide for the Care and Use of Laboratory Animals. A protocol was prepared before the study without registration. C57BL/6 mice were purchased from the Experimental Animal Center of Sun Yat-sen University, and kept under controlled environmental conditions (constant laminar airflow, 20–23 °C, 40–60% relative humidity and 12/12-h light/dark cycle). The vasodilation study was performed as described in our previous study (18). Briefly, 8-week-old male C57BL/6 mice were intravenously injected with 30 µg of Exos in 100 µL of PBS. After 24 h, the mice were anesthetized by pentobarbital sodium (50 mg/kg), and the thoracic aortas were isolated. The vascular rings with a diameter of 3 mm were tailored and then connected to an isometric force transducer (DMT 620M, Danish Myo Technology A/S, Denmark). The aortic rings were suspended in organ chambers filled with Krebs-solution and a gas mixture (95% oxygen and 5% carbon dioxide) was continuously supplied at 37 °C. The aortic rings were equilibrated for 60 min, and the solution was replaced every 15 min. The aortic rings were pre-constricted with 10−5 mol/L phenylephrine (P6126-5G, Sigma) and endothelium-dependent vasodilation was determined by acetylcholine (Ach, 10−9–10−5 mol/L; A6625-25G, Sigma) with or without NG-nitro-L-arginine methyl ester (100 µmol/L; N5751-1G, Sigma). Endothelium-independent vasodilation was assessed using sodium nitroprusside (10−9–10−5 mol/L; 228710-5G, Sigma).
Exosomal miRNA expression profiling
Total RNA was isolated using TRIzol reagent (Thermo Fisher Scientific, MA, USA). RNA concentration was measured using a Qubit® RNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, CA, USA), and RNA degradation and contamination were monitored on 1% agarose gels. After RNA quantification and qualification, 1 µg total RNA per sample was used as input material for the small RNA library. Sequencing libraries were generated using NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (NEB, USA) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample and the library quality was then assessed on the Agilent Bioanalyzer 2100 system using DNA High Sensitivity Chips. After cluster generation, the library preparations were sequenced on the Illumina NovaSeq 6000 platform, and 50 bp single-end reads were generated. Next, clean data (clean reads) were obtained by removing reads containing ploy N, with 5’ adapter contaminants, without 3’ adapter or the insert tag, containing ploy A or T or G or C and low-quality reads from the raw data. At the same time, Q20, Q30, and the guanine (G) and cytosine (C) content of the raw data were calculated. A certain range of length from the clean reads was chosen to do all the downstream analyses. miRNA expression levels were estimated by transcript per million, and a differential expression analysis between the two groups (three biological replicates) was performed using the DESeq (v1.22.1).
Target genes prediction and bioinformatics analysis
Potential target genes were predicted from the miRDB and miRBase databases. The Database for Annotation, Visualization, and Integrated Discovery was used to investigate the functional annotation of the target genes. A Gene Ontology (GO) analysis was performed to elaborate on the biological process, and a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed to explore the relevant signal pathway, and the networks were constructed on the Cytoscape platform (v3.8.2) (20). Protein-protein interaction (PPI) networks were identified using Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (21). A P value <0.05 was considered statistically significant.
Statistical analysis
The statistical analysis was conducted using SPSS 20.0 software (SPSS Inc., Chicago, IL, USA), and the graphs were plotted by GraphPad Prism (Version 7.0, San Diego, USA). The data are presented as mean ± standard error of the mean (SEM). For continuous variables with normal distributions, comparisons between two groups were performed with independent t-tests. For continuous variables with non-normal distributions, comparisons between two groups were performed with Wilcoxon rank-sum tests. A P value <0.05 was considered statistically significant.
Results
Isolation and characterization of Exos from hepatocyte culture supernatant
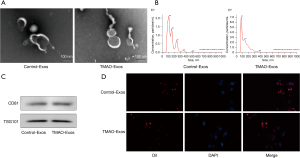
Nanovesicles with diameters of around 100 nm were isolated and purified from the hepatocyte culture supernatant, which were visualized under an electron microscope (see Figure 1A). The size distribution of these nanovesicles were further identified by nanoparticle tracking analysis (see Figure 1B). The expressions of the exosomal identity markers of CD81 and TSG101 were detected in Control-Exos and TMAO-Exos (see Figure 1C). The above results indicated that Exos have been successfully isolated and verified. Furthermore, the DiI-labelled Exos could be taken up by HAECs (see Figure 1D).
TMAO-Exos promoted inflammatory gene expression
Since TMAO has been recognized as a novel proinflammatory factor (5), we speculated that Exos derived from TMAO-stimulated hepatocytes may be involved with inflammation. HAECs were treated with Control-Exos and TMAO-Exos for 48 h and the mRNA expressions of inflammatory markers were detected. As Figure 2 shows, TMAO-Exos significantly increased the mRNA levels of IL-6, MCP-1, and TNF-α.

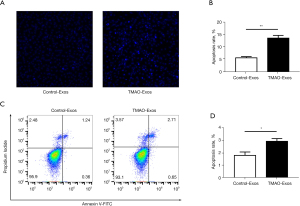
TMAO-Exos induced cell apoptosis and inhibited cell migration
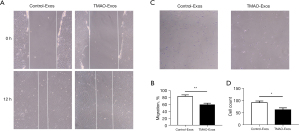
Next, we evaluated the effects of TMAO-Exos on cell apoptosis and migration. As Figure 3 shows, when exposed to Exos for 48 h, cell apoptosis was promoted by TMAO-Exos, which was detected by Hoechst staining (see Figure 3A,3B) and flow cytometry assay (see Figure 3C,3D). In Hoechst staining, apoptotic cells were characterized by a typical phenotype of chromatin condensation and fragmentation (22). Additionally, cell migration was determined to be inhibited by TMAO-Exos in scratch assay (see Figure 4A,4B) and transwell assay (see Figure 4C,4D).
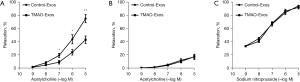
TMAO-Exos inhibited endothelium-dependent vasodilation
Then, we explored the effects of these Exos on endothelium-dependent vasodilation. It was showed that TMAO-Exos significantly reduced acetylcholine-dependent vasodilation (see Figure 5A). To clarify the role of endothelial nitric oxide synthase (eNOS) in this process, the vessels were pretreated with NG-nitro-L-arginine methyl ester (an eNOS inhibitor) for 30 min. As Figure 5B shows, the vasodilation was further inhibited and the differences between the two groups disappeared, indicating that eNOS played a role in mediating the inhibitory effects of TMAO-Exos on vasodilation. Furthermore, it was found that endothelium-independent vasodilation was not affected by the Exos (see Figure 5C).
The expression profiles of miRNAs in Exos, the prediction of miRNA-mRNA network, and bioinformatics analysis
An RNA-sequencing strategy was adopted to identify the differentially expressed miRNAs between the Control-Exos and TMAO-Exos groups. The miRNAs with a P<0.05 were visualized on a heatmap (see Figure 6A). Compared to the TMAO-free group, a total of 17 miRNAs changed significantly [| log2 (fold change) | >1; P<0.05] when exposed to TMAO (of these 17 miRNAs, 8 were up-regulated, and 9 were down-regulated; see Table 3). Genes labelled with a purely numerical code represented newly predicted miRNAs, and it is worth noting that 6 of them were down-regulated, but their functions remain unknown. It is well known that miRNA play a vital role in negatively regulating gene expression by destabilizing or inhibiting the translation of the target mRNA (23). Thus, the known miRNAs and another unknown with the highest expression were selected from Table 3 for the next analysis.
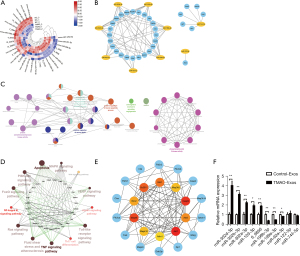
Table 3
| Type | #ID | Control-Exos_mean | TMAO-Exos_mean | Fold change_Log2 | P value |
|---|---|---|---|---|---|
| Up | 6_9856 | 53,136.4 | 114,742.9 | 1.110617915 | 4.24E−36 |
| 7_11542 | 44.3 | 660.1 | 3.867285559 | 0.042018456 | |
| miR-103-3p | 1,545.3 | 3,120.6 | 1.013465437 | 0.017122205 | |
| miR-302a-3p | 0 | 467 | 8.87036472 | 0.035026562 | |
| miR-302b-3p | 0 | 3,401.8 | 11.73250664 | 8.55E−12 | |
| miR-302d-3p | 0 | 766 | 9.583082768 | 0.003244933 | |
| miR-744-5p | 3,541.9 | 7,435.2 | 1.069634827 | 0.000621541 | |
| miR-92a-3p | 633.3 | 2,048.6 | 1.692105134 | 0.001897612 | |
| Down | 12_19875 | 3,430.1 | 1,407.5 | −1.28451161 | 0.005508287 |
| 15_23079 | 2,314.9 | 479.1 | −2.270166118 | 8.33E−05 | |
| 18_26561 | 492.8 | 54.4 | −3.155968955 | 0.023602876 | |
| 2_2684 | 513.3 | 37.4 | −3.743431938 | 0.014180519 | |
| 4_5543 | 443.1 | 17.1 | −4.616822969 | 0.032070247 | |
| 9_14879 | 1,712.3 | 456.2 | −1.905880482 | 0.001012913 | |
| miR-122-5p | 2,676.9 | 658.7 | −2.021220078 | 0.000116753 | |
| miR-199a-3p | 672.1 | 0 | −9.394677046 | 0.000582822 | |
| miR-199b-3p | 529 | 0 | −9.049848549 | 0.003405104 |
#, genes labelled with a purely numerical code represented newly predicted miRNAs. Exos, exosomes; TMAO, trimethylamine-N-oxide.
Thirty-one candidates predicted to interact with the differentially expressed miRNAs were selected, and miRNA-mRNA networks were built using Cytoscape software to show the complex interactions (see Figure 6B). The GO analysis revealed that changes in the biological processes of the predicted target genes were significantly enriched in the positive regulation of the apoptotic signaling pathway, the positive regulation of I-kappaB kinase/NF-kappaB signaling, and the response to endoplasmic reticulum stress. The interacting networks among these biological processes were constructed using ClueGO of Cytoscape (see Figure 6C). The KEGG analysis showed that these potential target genes were strongly associated with the signal pathways that were pivotal in regulating inflammation and endothelial function, such as NF-κB signaling pathway, TNF signaling pathway, and apoptosis, and the interactions among these signal pathways were constructed using ClueGO (see Figure 6D). Moreover, MCL1 apoptosis regulator (Mcl1), Bcl2l11, baculoviral IAP repeat containing 2 (Birc2), mitogen-activated protein kinase 8 (Mapk8), mitogen-activated protein kinase 10 (Mapk10), apoptosis inducing factor mitochondria associated 1 (Aifm1), AKT serine/threonine kinase 1 (Akt1), AKT serine/threonine kinase 3 (Akt3), caspase 9 (Casp9), and mitogen-activated protein kinase kinase kinase 5 (Map3k5) were identified as the top 10 hub genes (see Figure 6E). Further, it was verified that, when compared to the Control-Exos, miR-302d-3p, miR-302b-3p, miR-302a-3p, miR-103-3p, and 6_9856 (prediction) were up-regulated, while miR-199b-3p, miR-199a-3p, and miR-92a-3p were down-regulated (see Figure 6F).
TMAO-Exos promoted the activation of NF-κB signaling
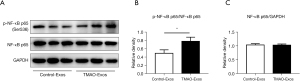
The GO and KEGG analyses of the predicted target genes of miRNAs indicated that TMAO-Exos may be involved in the NF-κB signaling pathway, therefore, we then investigated the effects of TMAO-Exos on NF-κB signaling in HAECs. It was found that, after incubation with Exos for 24 h, TMAO-Exos notably enhanced the expression of p-NF-κB p65 (see Figure 7A,7B), but had no effect on NF-κB p65 (see Figure 7A,7C).
Discussion
In the current research, TMAO was found to directly stimulate hepatocytes to release Exos that could be taken up by HAECs, thus promoting the expressions of inflammatory markers and cell apoptosis, impairing cell migration and inhibiting endothelium-dependent vasodilation. Next, we characterized the miRNAs contained in the Exos, and further revealed that these differentially expressed miRNAs were predicted to target the potential genes that involved in inflammation and endothelial function. Finally, we verified that NF-κB signaling was activated by TMAO-Exos.
In recent years, many studies have shown that intestinal flora and metabolites exert regulatory effects on atherosclerosis by inhibiting or accelerating the disease process (24). It has been confirmed that dietary choline and phosphatidylcholine can be metabolized into trimethylamine (TMA) in intestinal microbiota, and TMA can then be further converted to TMAO under the catalysis of the flavin monooxygenase-3 in the liver (14). TMAO has been proven to be a novel independent risk factor for atherosclerosis and serious cardiovascular events (14,25,26). Further, numerous studies have shown that TMAO promotes atherosclerosis by boosting inflammatory activation and impairing endothelial function both in vitro and in vivo (3,5,27). Notably, increased blood TMAO levels in low-density lipoprotein receptor-deficient mice contributed to the activation of NF-κB pathway and promoted the expressions of inflammatory markers (5). Additionally, elevated plasma TMAO levels in rats were found to inhibit eNOS expression and enhance the production of inflammatory cytokine and superoxide, resulting in senescence-related endothelial dysfunction (27). In-vitro experiments have shown that TMAO induces inflammation and endothelial dysfunction by suppressing the activity of eNOS, provoking oxidative stress and activating inflammasome (3). However, the effective concentrations of TMAO on VECs in vitro were much higher than the actual levels in the body (3,5,7,14,25-28). This may be due to the complexity of the internal environment. Conversely, it could suggest that understandings of the molecular mechanisms of TMAO are incomplete, and indirect factors contributing to inflammation and endothelial dysfunction cannot be excluded. As TMAO is produced in the liver, hepatocytes may be the first potential target for TMAO. Evidence from recent related studies support our idea that a relationship exists between TMAO and hepatocytes. Specifically, Chen et al. recently found that a physiological concentration of TMAO directly binds to hepatic protein kinase R-like endoplasmic reticulum kinase (PERK), thereby activating the unfolded protein response and promoting metabolic dysfunction (15). Another study used animal models of atherosclerosis and found that hepatic miR-146a-5p expression was associated with blood TMAO (29).
Recent studies have shown that Exos, including those derived from hepatocytes, are involved in atherosclerosis by regulating inflammation and endothelial function (12,30). Additionally, Hirsova et al. found that primary hepatocytes and Huh7 cells stimulated with lipid released more extracellular vesicles (which possessed the characteristics of Exos), thereby enhancing the expressions of IL-1β and IL-6 in macrophages via tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) contained in these vesicles (13). However, to the best of our knowledge, no studies have defined the relationship between hepatocyte-derived Exos and the detrimental effects of TMAO on VECs. In the present study, we found that TMAO can stimulate hepatocytes to release Exos, and what's more, these TMAO-activated Exos play a role in enhancing the expressions of IL-6, MCP-1 and TNF-α, inducing cell apoptosis, inhibiting cell migration, and impairing endothelium-dependent vasodilation.
IL-6, MCP-1, and TNF-α have been identified as key inflammatory factors that play pivotal roles in the regulation of inflammation, endothelial dysfunction, and atherosclerosis. IL-6 has been confirmed to be an independent predictor of plaque progression (31). In hypertensive patients with coronary artery disease, blood MCP-1 levels were elevated and were related to the degree of endothelial damage (32), and MCP-1 secretion from VECs was involved in atherosclerosis (33). The role of TNF-α in impairing endothelial function has also been established (34). Additionally, increased cell apoptosis and decreased cell migration have been recognized as key features of endothelial injury (18). Endothelial dysfunction is the initial event of atherogenesis, which plays a key role in the development and progression of atherosclerosis (35). Once the endothelial barrier is impaired, lipid infiltration and monocyte recruitment occur, then foam cells form and the inflammatory response is continuedly activated (2,36). Additionally, injured VECs also trigger a series of cellular and inflammatory responses (35).
The miRNAs carried by Exos are promising biomarkers for the early detection of cardiovascular disease (37). Additionally, these exosomal miRNAs can be actively absorbed by both neighboring and distal cells and thus exert regulatory roles in the disease process (37). Recent research has shown that steatotic hepatocyte-derived Exos could transfer miR-1 to VECs and contribute to inflammatory activation and atherogenesis by inhibiting Kruppel-like factor 4 expression and activating the NF-κB pathway (12). Zheng et al. found that Exos secreted from vascular smooth muscle cells (VSMCs) were able to mediate the communication between VSMCs and VECs by transferring Kruppel-like factor 5-induced miR-155 to VECs, thereby damaging endothelial function and accelerating atherogenesis progression (38). In our study, miRNAs carried by the TMAO-Exos were quite different to those in the TMAO-free group, including several well-recognized miRNAs, such as miR-302d-3p, miR-103-3p, miR-199a-3p, and miR-92a-3p. The bioinformatics analysis showed that the potential target genes for these miRNAs were supposed to be involved in the signal pathways related to inflammation and endothelial function. Further, miRNAs, such as miR-199a and miR-199b, have been shown to regulate inflammation by directly inhibiting NF-κB signaling (39-41). There is extensive evidence that the NF-κB signaling pathway, TNF signaling pathway, apoptosis signaling pathway, toll-like receptor signaling pathway, and forkhead box, sub-group O signaling pathway play crucial roles in inflammation, endothelial function, and atherosclerosis (34,42-46). The NF-κB signaling pathway is at the crossroads of inflammation and atherogenesis (45), and the specific NF-κB inhibition of endothelial cells has been shown to reduce inflammatory gene expression, such as IL-6 and TNF, and protect apolipoprotein E-deficient mice from atherosclerosis (42). In addition, recent studies have shown that Exos promote atherosclerosis through the NF-κB pathway (30,47). In our study, we found that NF-κB p65 could be activated by TMAO-Exos. The relationships between the NF-κB pathway and the aforementioned inflammatory cytokines were close. IL-6 and MCP-1 contain the binding elements for NF-κB, which are critical in the transcriptional induction of IL-6 and MCP-1 genes, and TNF-α served as one of the NF-kB activating factors, inducing IL-6 and MCP-1 transcription intensely (48,49).
Overall, the results of the present study provide evidence that hepatocyte-derived Exos and miRNAs carried by TMAO-Exos mediate the detrimental effects of TMAO on VECs. However, understandings of how TMAO stimulates the secretion of Exos from hepatocytes and how miRNAs are selectively assembled on these Exos are limited. Future research designed to investigate the precise mechanisms by which exosomal miRNAs interact with their target genes to irritate inflammation and endothelial dysfunction would deepen our understanding of the roles of TMAO in VECs.
Conclusions
In the current study, we found that Exos derived from TMAO-stimulated hepatocytes promoted the expressions of inflammatory markers and endothelial dysfunction, at least in part, by NF-κB signaling. Additionally, the differentially expressed miRNAs carried by TMAO-Exos appeared to mediate the adverse effects on VECs by regulating the expressions of their target genes. Our findings provide novel insights into the interactions between liver and VECs in the atherogenic process orchestrated by TMAO, and suggest that targeting the cellular crosstalk may provide an effective approach for restraining the detrimental effects induced by TMAO.
Acknowledgments
Funding: This work was supported by the National Key Research and Development Program of China (No. 2018YFC1002600, No. 2020YFC2008005), Guangdong Peak Project (No. DFJH2019), Science and Technology Projects in Guangzhou (No. 202102021149, No. 202002020030), Postdoctoral Scientific Research Start-up Fund Project of Guangdong Provincial People’s Hospital (BY012021052), National Nature Science Foundation of China (No. 82100451).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-5043
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-5043
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-5043). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The animal experiments were performed under a project license (No. S2020-168) granted by the ethics committees for animal experiments of Guangzhou Medical University, in compliance with the National Research Council's Guide for the Care and Use of Laboratory Animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Roth GA, Mensah GA, Johnson CO, et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990-2019: Update From the GBD 2019 Study. J Am Coll Cardiol 2020;76:2982-3021. [Crossref] [PubMed]
- Spann NJ, Garmire LX, McDonald JG, et al. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell 2012;151:138-52. [Crossref] [PubMed]
- Sun X, Jiao X, Ma Y, et al. Trimethylamine N-oxide induces inflammation and endothelial dysfunction in human umbilical vein endothelial cells via activating ROS-TXNIP-NLRP3 inflammasome. Biochem Biophys Res Commun 2016;481:63-70. [Crossref] [PubMed]
- Wu P, Chen J, Chen J, et al. Trimethylamine N-oxide promotes apoE-/- mice atherosclerosis by inducing vascular endothelial cell pyroptosis via the SDHB/ROS pathway. J Cell Physiol 2020;235:6582-91. [Crossref] [PubMed]
- Seldin MM, Meng Y, Qi H, et al. Trimethylamine N-Oxide Promotes Vascular Inflammation Through Signaling of Mitogen-Activated Protein Kinase and Nuclear Factor-κB. J Am Heart Assoc 2016;5:002767 [Crossref] [PubMed]
- Liu Y, Dai M. Trimethylamine N-Oxide Generated by the Gut Microbiota Is Associated with Vascular Inflammation: New Insights into Atherosclerosis. Mediators Inflamm 2020;2020:4634172 [Crossref] [PubMed]
- Singh GB, Zhang Y, Boini KM, et al. High Mobility Group Box 1 Mediates TMAO-Induced Endothelial Dysfunction. Int J Mol Sci 2019;20:3570. [Crossref] [PubMed]
- Kourembanas S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol 2015;77:13-27. [Crossref] [PubMed]
- Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002;2:569-79. [Crossref] [PubMed]
- Xu Z, Zhou X, Wu J, et al. Mesenchymal stem cell-derived exosomes carrying microRNA-150 suppresses the proliferation and migration of osteosarcoma cells via targeting IGF2BP1. Transl Cancer Res 2020;9:5323-35. [Crossref]
- Momen-Heravi F, Bala S, Kodys K, et al. Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Sci Rep 2015;5:9991. [Crossref] [PubMed]
- Jiang F, Chen Q, Wang W, et al. Hepatocyte-derived extracellular vesicles promote endothelial inflammation and atherogenesis via microRNA-1. J Hepatol 2020;72:156-66. [Crossref] [PubMed]
- Hirsova P, Ibrahim SH, Krishnan A, et al. Lipid-Induced Signaling Causes Release of Inflammatory Extracellular Vesicles From Hepatocytes. Gastroenterology 2016;150:956-67. [Crossref] [PubMed]
- Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011;472:57-63. [Crossref] [PubMed]
- Chen S, Henderson A, Petriello MC, et al. Trimethylamine N-Oxide Binds and Activates PERK to Promote Metabolic Dysfunction. Cell Metab 2019;30:1141-1151.e5. [Crossref] [PubMed]
- Nagarajan SR, Paul-Heng M, Krycer JR, et al. Lipid and glucose metabolism in hepatocyte cell lines and primary mouse hepatocytes: a comprehensive resource for in vitro studies of hepatic metabolism. Am J Physiol Endocrinol Metab 2019;316:E578-89. [Crossref] [PubMed]
- Wu JC, Merlino G, Fausto N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc Natl Acad Sci U S A 1994;91:674-8. [Crossref] [PubMed]
- Ou ZJ, Chen J, Dai WP, et al. 25-Hydroxycholesterol impairs endothelial function and vasodilation by uncoupling and inhibiting endothelial nitric oxide synthase. Am J Physiol Endocrinol Metab 2016;311:E781-90. [Crossref] [PubMed]
- Zhou L, Liu X, Wang ZQ, et al. Simvastatin Treatment Protects Myocardium in Noncoronary Artery Cardiac Surgery by Inhibiting Apoptosis Through miR-15a-5p Targeting. J Cardiovasc Pharmacol 2018;72:176-85. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Kumar S, Kasseckert S, Kostin S, et al. Ischemic acidosis causes apoptosis in coronary endothelial cells through activation of caspase-12. Cardiovasc Res 2007;73:172-80. [Crossref] [PubMed]
- De Rosa S, Curcio A, Indolfi C. Emerging role of microRNAs in cardiovascular diseases. Circ J 2014;78:567-75. [Crossref] [PubMed]
- Chistiakov DA, Bobryshev YV, Kozarov E, et al. Role of gut microbiota in the modulation of atherosclerosis-associated immune response. Front Microbiol 2015;6:671. [Crossref] [PubMed]
- Koeth RA, Wang Z, Levison BS, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 2013;19:576-85. [Crossref] [PubMed]
- Tang WH, Wang Z, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 2013;368:1575-84. [Crossref] [PubMed]
- Li T, Chen Y, Gua C, et al. Elevated Circulating Trimethylamine N-Oxide Levels Contribute to Endothelial Dysfunction in Aged Rats through Vascular Inflammation and Oxidative Stress. Front Physiol 2017;8:350. [Crossref] [PubMed]
- Brunt VE, Gioscia-Ryan RA, Casso AG, et al. Trimethylamine-N-Oxide Promotes Age-Related Vascular Oxidative Stress and Endothelial Dysfunction in Mice and Healthy Humans. Hypertension 2020;76:101-12. [Crossref] [PubMed]
- Coffey AR, Kanke M, Smallwood TL, et al. microRNA-146a-5p association with the cardiometabolic disease risk factor TMAO. Physiol Genomics 2019;51:59-71. [Crossref] [PubMed]
- Gao W, Liu H, Yuan J, et al. Exosomes derived from mature dendritic cells increase endothelial inflammation and atherosclerosis via membrane TNF-α mediated NF-κB pathway. J Cell Mol Med 2016;20:2318-27. [Crossref] [PubMed]
- Eltoft A, Arntzen KA, Wilsgaard T, et al. Interleukin-6 is an independent predictor of progressive atherosclerosis in the carotid artery: The tromso study. Atherosclerosis 2018;271:1-8. [Crossref] [PubMed]
- Tucci M, Quatraro C, Frassanito MA, et al. Deregulated expression of monocyte chemoattractant protein-1 (MCP-1) in arterial hypertension: role in endothelial inflammation and atheromasia. J Hypertens 2006;24:1307-18. [Crossref] [PubMed]
- Dje N'Guessan P, Riediger F, Vardarova K, et al. Statins control oxidized LDL-mediated histone modifications and gene expression in cultured human endothelial cells. Arterioscler Thromb Vasc Biol 2009;29:380-6. [Crossref] [PubMed]
- Deng X, Chu X, Wang P, et al. MicroRNA-29a-3p Reduces TNFα-Induced Endothelial Dysfunction by Targeting Tumor Necrosis Factor Receptor 1. Mol Ther Nucleic Acids 2019;18:903-15. [Crossref] [PubMed]
- Gimbrone MA Jr, García-Cardeña G. Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ Res 2016;118:620-36. [Crossref] [PubMed]
- Aryal B, Suárez Y. Non-coding RNA regulation of endothelial and macrophage functions during atherosclerosis. Vascul Pharmacol 2019;114:64-75. [Crossref] [PubMed]
- Aghabozorgi AS, Ahangari N, Eftekhaari TE, et al. Circulating exosomal miRNAs in cardiovascular disease pathogenesis: New emerging hopes. J Cell Physiol 2019;234:21796-809. [Crossref] [PubMed]
- Zheng B, Yin WN, Suzuki T, et al. Exosome-Mediated miR-155 Transfer from Smooth Muscle Cells to Endothelial Cells Induces Endothelial Injury and Promotes Atherosclerosis. Mol Ther 2017;25:1279-94. [Crossref] [PubMed]
- Dai L, Gu L, Di W. MiR-199a attenuates endometrial stromal cell invasiveness through suppression of the IKKβ/NF-κB pathway and reduced interleukin-8 expression. Mol Hum Reprod 2012;18:136-45. [Crossref] [PubMed]
- Zhang R, Qin L, Shi J. MicroRNA-199a-3p suppresses high glucose-induced apoptosis and inflammation by regulating the IKKβ/NF-κB signaling pathway in renal tubular epithelial cells. Int J Mol Med 2020;46:2161-71. [Crossref] [PubMed]
- Zhu L, Xu H, Lv W, et al. miR-199b-5p Regulates Immune-Mediated Allograft Rejection after Lung Transplantation Through the GSK3β and NF-κB Pathways. Inflammation 2018;41:1524-35. [Crossref] [PubMed]
- Gareus R, Kotsaki E, Xanthoulea S, et al. Endothelial cell-specific NF-kappaB inhibition protects mice from atherosclerosis. Cell Metab 2008;8:372-83. [Crossref] [PubMed]
- Heo KS, Lee H, Nigro P, et al. PKCζ mediates disturbed flow-induced endothelial apoptosis via p53 SUMOylation. J Cell Biol 2011;193:867-84. [Crossref] [PubMed]
- Menghini R, Casagrande V, Iuliani G, et al. Metabolic aspects of cardiovascular diseases: Is FoxO1 a player or a target? Int J Biochem Cell Biol 2020;118:105659 [Crossref] [PubMed]
- Pateras I, Giaginis C, Tsigris C, et al. NF-κB signaling at the crossroads of inflammation and atherogenesis: searching for new therapeutic links. Expert Opin Ther Targets 2014;18:1089-101. [Crossref] [PubMed]
- Polykratis A, van Loo G, Xanthoulea S, et al. Conditional targeting of tumor necrosis factor receptor-associated factor 6 reveals opposing functions of Toll-like receptor signaling in endothelial and myeloid cells in a mouse model of atherosclerosis. Circulation 2012;126:1739-51. [Crossref] [PubMed]
- Xu L, Geng T, Zang G, et al. Exosome derived from CD137-modified endothelial cells regulates the Th17 responses in atherosclerosis. J Cell Mol Med 2020;24:4659-67. [Crossref] [PubMed]
- Murakami M, Kamimura D, Hirano T. Pleiotropy and Specificity: Insights from the Interleukin 6 Family of Cytokines. Immunity 2019;50:812-31. [Crossref] [PubMed]
- Ping D, Boekhoudt G, Zhang F, et al. Sp1 binding is critical for promoter assembly and activation of the MCP-1 gene by tumor necrosis factor. J Biol Chem 2000;275:1708-14. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


