
PDGF-BB promotes vascular smooth muscle cell migration by enhancing Pim-1 expression via inhibiting miR-214
Introduction
Cardiovascular diseases (CVD) are a group of diseases which mainly the vessels and organ of the heart (1). Atherosclerosis, a universal type of CVD involving a process of accumulation of fatty and/or fibrous material in the innermost layer of arteries, has resulted in significant morbidity and mortality worldwide (2). Atherosclerosis is an inexplicable and multistage process which blocks blood flow to the heart and brain, resulting in the initiation of many other CVD (3). Many studies have shown that the anomalous cell migration and proliferation of vascular smooth muscle cells (VSMC) may be a critical element in the progression of atherosclerosis (4,5).
MicroRNAs (miRNAs) belong to small non-coding RNA family, which universally function by regulating the expression of downstream genes (6). It has also been shown that miRNAs are involved in physiological processes of the cardiovascular system, including regulation of development of the heart, controlling cardiac function, and engaging in the apoptosis of cardiomyocytes (7,8). Several studies have also illustrated that miRNAs participate in the development of coronary atherosclerosis (9,10). These findings suggest that miRNAs could be potential novel biomarkers for the early diagnosis and prognosis, as well as therapeutic targets for CVD. It has been reported that miR-214 expression is abnormally regulated in several diseases (11,12); however, its function in CVD development and the underlying mechanisms still remain unclear. The platelet-derived growth factor/platelet-derived growth factor receptor (PDGF/PDGFR) pathway plays an important physiological role in normal growth, development, tumorigenesis, and is closely involved in the development of CVD such as atherosclerosis (13). However, despite the large volume of research delineating the function and underlying mechanisms of the PDGF/PDGFR pathway in the process of atherosclerosis, whether miR-214 is involved remains unknown.
In the current study, we found that PDGF-BB could enhance smooth muscle cell migration by promoting epithelial-mesenchymal transition (EMT). And for the first time, we identified PDGF-BB could suppress the expression of miR-214 and subsequently elevate the expression of Pim-1, a target gene of miR-214. Rescue experiments confirmed that the upregulation of Pim-1expression by miR-214 inhibition is indispensable for the role of PDGF-BB on A7r5 cells. Moreover, both PDGF-BB and Pim-1 could promote the activation of AKT, ERK, and STAT3. All these findings suggested that the PDGF/miR-214/Pim-1 axis could be a therapeutic target for coronary atherosclerotic heart disease. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/atm-21-5638).
Methods
Cell culture
The rat smooth muscle cell line A7r5 (CRL-1444) was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were cultured in Dulbecco’s modified Eagle medium (DMEM; Hyclone, Logan, UT, USA) plus 10% fetal bovine serum (FBS; Hyclone, USA), 100 U/mL of penicillin (Gibco BRL, Rockville, MD, USA) and 100 µg/mL of streptomycin (Gibco BRL, USA) in a humidified incubator with 5% (v/v) CO2 at 37 °C.
In vitro migration assays
The Transwell system (Corning Life Sciences, Tewksbury, MA, USA) was used to determine the cell migration potential of A7r5 cells. A total of 20,000 cells in serum-free DMEM were seeded into the upper chambers, and the lower chamber was filled with 0.5 mL DMEM medium with 10% fetal bovine serum (FBS). Cells were allowed to migrate for 24 h in a culture incubator. After incubation, the cells were fixed with 4% paraformaldehyde for 15 min at room temperature, and the remaining cells on the upper membrane were removed, followed by 10 min staining with 0.5% crystal violet at room temperature.
Western blotting
Cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), proteins were separated and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Burlington, MA, USA) followed by blocking with 5% non-fat milk and primary antibodies incubation. The primary antibodies were used as follows: Pim-1 (CST-54523, 1:1,000 dilution), E-cadherin (proteintech-20874, 1:1,000 dilution; Proteintech, Rosemont, IL, USA), N-cadherin (proteitech-22018, 1:1,000 dilution; Proteintech, USA), vimentin (proteitech-10366, 1:1,000 dilution; Proteintech, USA), STAT3 (proteitech-10253, 1:1,000 dilution; Proteintech, USA), p-STAT3 [CST-9145, 1:1,000 dilution; Cell Signaling Technology (CST), Danvers, MA, USA], ERK (proteitech-20874, 1:1,000 dilution; Proteintech, USA), p-ERK (CST-4370, 1:1,000 dilution; CST, USA), AKT (CST-4685, 1:1,000 dilution; CST, USA), p-AKT (CST-4060, 1:1,000 dilution; CST, USA), and β-actin (Sigma, 1:5,000 dilution; Sigma-Aldrich, St. Louis, MO, USA). All primary antibodies were incubated at 4 °C overnight, and the horseradish peroxidase (HRP)-conjugated secondary antibody was incubated with 1:5,000 dilution for 2 h at room temperature.
RNA isolation and quantitative real-time polymerase chain reaction
Total RNA was isolated from samples of treated cells using the Trizol (Invitrogen, USA) and converted to cDNA using the PrimeScript RT Reagent Kit (TaKaRa, Osaka, Japan). The TaqMan microRNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) was used to synthesize miRNA to cDNA. The real-time PCR assay was performed using SYBR green PCR master mix (Applied Biosystems, USA) using the 2-△△Ct method. The primer sequences used were as follows: miR-214 (RT: 5'-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATA CGACCCTGCT-3'; forward: 5'-AGGGTCCGAGGTATTCG-3'; reverse: 5'-AATGGATTTT TG-3'), U6 (forward: 5'-CTCGCTTCGGCAGCACA-3'; reverse 5'-AACGCTTCACGAATTT GCGT-3'), Pim-1 (forward: 5'-GATGGGACCCGAGTGTAT-3'; reverse 5’-GGATGGTTCT GGATTTCT-3'), β-actin (forward: 5'-AGCAGTTGTAGCTACCCGCCCA-3'; reverse 5'-GGCGGGCACGTTG AAGGTCT-3').
miR-214 mimics transfection
The miR-214 mimics and negative control (NC) mimic oligonucleotides were synthesized by Ribobio (Guangzhou, China). A total of 1×105 cells were seeded in a 6-well plate and incubated overnight to allow to attachment. The transfection was performed using Lipofectamine 2000 (Invitrogen, USA) and the expression was validated by real-time PCR.
Cell cycle analysis by flow cytometry
Cell cycle distribution was determined by flow cytometry (Attune® NxT, Thermo Fisher, Waltham, MA, USA) after propidium oxide staining with a CycleTest Plus DNA Reagent Kit (Becton, Dickinson and Co. Biosciences, Franklin Lakes, NJ, USA). The numbers of cells in each phase were assessed. The percentages of cells at G1, S, and G2/M were calculated by the FlowJo software (Flowjo, LLC, Ashland, OR, USA).
Pim-1 overexpression plasmid construction and transfection
The ORF sequences of human Pim-1 mRNA were cloned into the overexpression vector pcDNA3.1. For transfection, Lipofectamine 2000 (Invitrogen, USA) was mixed with the plasmids for 20 min at room temperature to form a complex according to manufacturer’s instructions. After incubation for 28 h at 37 °C, the overexpression efficiency was determined by western blotting.
Luciferase assay
The A7r5 cells were seeded in 24-well plates overnight and treated with PDGF-BB for another 24 h. Next, cells were transfected with 1.2 µg PGL3 luciferase reporter with wild type Pim-1 3’UTR (WT) or mutant Pim-1 3’UTR (MT) plasmids using Lipofectamine 2000 reagent and incubated for another 24 h. A PGL3 empty vector was used as a negative control and pRL-CMV plasmid was co-transfected as a positive control to normalize transfection efficiency. Then, Dual-Luciferase Reporter Assay System kit (Promega) was used to determine the luciferase activity using a microplate reader (Promega, Madison, WI, USA).
Statistical analysis
All data were expressed as the mean ± SD. Statistical analysis was performed using GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA). A value of P<0.05 was considered statistically significant.
Results
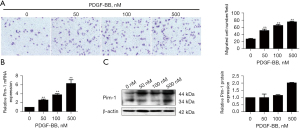
PDGF-BB enhanced cell migration of A7R5 cells and promoted Pim-1 expression
In this study, PDGF-BB was used to stimulate smooth muscle cell A7r5, and its dose and time for cellular treatment were used as previous (14). Our data indicated that the cell migration of A7r5 cells was significantly increased by PDGF-BB in a dose-dependent manner (Figure 1A). The expression levels of Pim-1 mRNA and protein in A7r5 cell were also up-regulated after treatment by PDGF-BB for 24 h (Figure 1B,1C). Taken together, these results suggested that up-regulation of Pim-1 by PDGF-BB might contribute to the cell migration of smooth muscle cells.
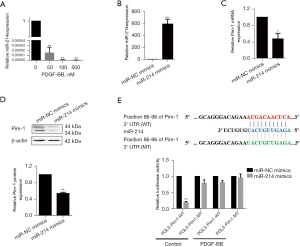
Pim-1 is a target of miR-214 and PDGF-BB effectively suppresses the expression of miR-214 in A7r5 cells
Although little is known about the role of miR-214 in CVD, our data indicated that PDGF-BB could effectively downregulate the expression of miR-214 in A7r5 cells (Figure 2A). We used 2 datasets of TargetScan (https://www.targetscan.org) and miRTarBase (https://www.mirtarbase.mbc.nctu.edu.tw) to search the potential targets of miR-214. Both datasets predicted Pim-1 as a potential target of miR-214. To investigate whether miR‑214 could regulate Pim-1 expression in A7r5 cells, miR‑214 mimics were used and the transfection efficiency was examined by quantitative polymerase chain reaction (qPCR). Our data revealed that miR‑214 was successfully expressed after transfection compared with NC group (Figure 2B). More importantly, the data from qRT-PCR and western blot indicated that both Pim-1 mRNA and protein levels were significantly reduced by the ectopic expression of miR‑214 in the A7r5 cells (Figure 2C,2D). Finally, luciferase assay showed that miR-214 obviously suppressed the luciferase activity of the plasmid with wild 3’UTR of Pim-1 mRNA, while failing to function on the plasmid with mutant 3’UTR of Pim-1 messenger RNA (mRNA). Furthermore, PDGF-BB effectively recovered the luciferase activity inhibited by miR-214 (Figure 2E). These results indicate that PDGF-BB could inhibit the expression of miR‑214, and subsequently suppress Pim-1 expression in A7r5 cells.
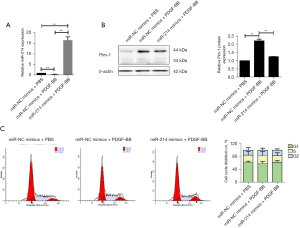
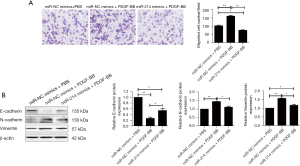
miR-214 inhibited PDGF-BB-induced cell migration in A7r5 cells by repressing EMT
To explore whether miR-214 was involved in the regulation of Pim-1 expression by PDGF-BB stimulation, the A7r5 cells were transfected with miR-214 mimics or NC mimics following PDGF-BB stimulation or control, and it was found that miR-214 were much more highly expressed after transfection, while PDGF-BB inhibited the expression of miR-214 (Figure 3A). Also, as shown in Figure 3B, the results showed that Pim-1 were highly expressed after PDGF-BB stimulation, while miR-214 mimic transfection decreased Pim-1 expression even under PDGF-BB treatment. Our data indicated that significant change was observed in the cell cycle distribution of A7r5 cells after PDGF-BB stimulation compared with control (Figure 3C). However, miR-214 significantly attenuated the enhanced cell migration of A7r5 cells induced by PDGF-BB (Figure 4A). More importantly, PDGF-BB effectively upregulated the expression of E-cadherin, while it downregulated the expression of N-cadherin and vimentin, and miR-214 mimics significantly partially reversed the role of PDGF-BB (Figure 4B). Taken together, PDGF-BB could promote A7r5 cells migration by promoting EMT via suppressing miR-214 expression at least partially.
The miR-214/Pim-1 axis mediated the cell migration induced by PDGF-BB in A7r5 cells
To further examine the function of the miR-214/Pim-1 axis in A7r5 cell migration stimulated by PDGF-BB, a Pim-1 overexpression plasmid was constructed. The A7r5 cells firstly were transfected with pcDNA3.1 or pcDNA-Pim-1 and the transfection efficiency was examined. It was observed that Pim-1 was highly expressed after transfection for 48 h (Figure 5A). Additionally, the qPCR results showed that the relative miR-214 expression was significantly decreased in A7r5 cells treated by PDGF-BB after co-transfecting with miR-214 mimics and pcDNA-Pim-1, compared with control groups, respectively (Figure 5B). Moreover, as shown in Figure 5C, PDGF-BB effectively induced the expression of Pim-1, while miR-214 alone not only suppressed the Pim-1 expression, but also successfully reversed the Pim-1 expression induced by PDGF-BB.

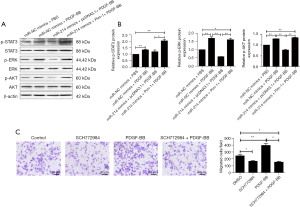
Our data further revealed that the cell migration of A7r5 cells were increased after PDGF-BB stimulation and was impaired after the transfection of miR-214 mimics in A7r5 cells treated by PDGF-BB (Figure 5D). Moreover, miR-214-mediated downregulation of the cell migration in A7r5 cells was effectively reversed by overexpression of Pim-1 (Figure 5D). Finally, several signaling pathways downstream of PDGF were also determined. As shown in Figure 6A,6B, the phosphorylation of AKT, ERK, and STAT3 was significantly enhanced by both PDGF-BB and Pim-1 overexpression, while weakened by miR-214 mimics. Moreover, a novel ERK inhibitor SCH772984 was used to confirm the role of PDGF-BB on cell migration of A7r5 cells. As shown in Figure 6C, SCH772984 not only repressed the cell migration potential of A7r5 cells, but also attenuated the role PDGF-BB.
Discussion
The main cause of CAD is atherosclerotic stenosis (15). In the pathological progression of atherosclerosis, vascular injuries result in the drive of various growth factors in VSMC (16). It has been shown that PDGF takes part in regulating the formation of blood vessel by influencing VSMC proliferation and migration (17,18). The PDGF family consists of 5 members including PDGF-AA, PDGF-BB, PDGFAB, PDGF-CC, and PDGF-DD (19). Among them, PDGF-BB is found to be most likely potential stimulant for switching the phenotypical VSMC from a retractable phenotype to a factitious one (13). Consequently, PDGF-BB is frequently employed as a stimulant to induce the phenotypical VSMC transition (20). In our study, PDGF-BB was also adopted to stimulate smooth muscle cell migration.
It has been identified that Pim-1, a serine/threonine protein kinase, plays a pivotal role in tumorigenesis (21). It has also been found to associate with vascular diseases by taking part in regulating VSMC proliferation (22,23). Moreover, an enhanced expression of Pim-1 in response to PDGF-BB stimulation was observed in A7r5 cells in this work, which was supported by a previous study (23). However, the underlying mechanism for how PDGF-BB regulates Pim-1 expression in A7r5 cells has remained unknown.
It has been reported that miRNAs are involved in progression and phenotypic transition in both cardiovascular cells and tissues (24). They also join in the development and progression of CVD (25,26). For instance, the downregulation of miR-145, miR-126, miR-155, and miR-17, while the up-regulation of miR-208 and miR-133 were observed in plasma and serum of the CAD group with drug treatment (27). In the present study, our results revealed that miR-214 was decreased in A7r5 cells stimulated by PDGF-BB. Upregulating miR-214 was shown to have negative effects on A7r5 cell migration stimulated by PDGF-BB, but there was no change in cell cycle distribution.
The datasets analysis demonstrated that Pim-1 was a potential target gene of miR-214, and several studies have also showed that miR-214 could target the expression of Pim-1 in some cell models such as mesenchymal stem cells and hepatocytes (28,29). Our data also revealed, for the first time (to the best of our knowledge) that miR-214 could negatively regulate Pim-1 expression in A7r5 cells by binding to the 3’UTR region. More importantly, enhanced migration activities of smooth muscle cells by PDGF-BB stimulation could be reversed by miR-214 overexpression, while the ectopic expression of Pim-1 also further effectively attenuated the inhibitory function of miR-214 in A7r5 cell migration. Therefore, all our finding in the present study clearly illustrated that miR-214 suppressed PDGF-BB-induced smooth muscle cell migration through targeting Pim-1.
Conclusions
In conclusion, our data demonstrated that PDGF-BB enhanced smooth muscle cell migration by suppressing miR-214 and promoting expression of its target gene Pim-1. Our findings suggest that PDFG/miR-214/Pim-1 axis could be a potential therapeutic target for coronary atherosclerotic heart disease.
Acknowledgments
Funding: This work was supported by funds from Zhejiang Provincial Natural Science Foundation of China/Exploration Program (LQ19H020010) and Yiwu Science and Technology Bureau (20-3-108).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-5638
Data Sharing Statement: Available athttps://dx.doi.org/10.21037/atm-21-5638
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-5638). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Libby P, Bornfeldt KE, Tall AR. Atherosclerosis: Successes, Surprises, and Future Challenges. Circ Res 2016;118:531-4. [Crossref] [PubMed]
- Libby P, Buring JE, Badimon L, et al. Atherosclerosis. Nat Rev Dis Primers 2019;5:56. [Crossref] [PubMed]
- Wang H, Nie ZY, Liu M, et al. Clinical characteristics of perivascular space and brain CT perfusion in stroke-free patients with intracranial and extracranial atherosclerosis of different extents. Ann Transl Med 2020;8:215. [Crossref] [PubMed]
- Zhao D, Li J, Xue C, et al. TL1A inhibits atherosclerosis in apoE-deficient mice by regulating the phenotype of vascular smooth muscle cells. J Biol Chem 2020;295:16314-27. [Crossref] [PubMed]
- Chong H, Wei Z, Na M, et al. The PGC-1α/NRF1/miR-378a axis protects vascular smooth muscle cells from FFA-induced proliferation, migration and inflammation in atherosclerosis. Atherosclerosis 2020;297:136-45. [Crossref] [PubMed]
- Sharma PC, Gupta A. MicroRNAs: potential biomarkers for diagnosis and prognosis of different cancers. Transl Cancer Res 2020;9:5798-818. [Crossref]
- Yang X, Du X, Ma K, et al. Circulating miRNAs Related to Long-term Adverse Cardiovascular Events in STEMI Patients: A Nested Case-Control Study. Can J Cardiol 2021;37:77-85. [Crossref] [PubMed]
- Devaux Y, Badimon L. CDR132L: another brick in the wall towards the use of miRNAs to treat cardiovascular disease. Eur Heart J 2021;42:202-4. [Crossref] [PubMed]
- Karunakaran D, Rayner KJ. Macrophage miRNAs in atherosclerosis. Biochim Biophys Acta 2016;1861:2087-93. [Crossref] [PubMed]
- Lu Y, Thavarajah T, Gu W, et al. Impact of miRNA in Atherosclerosis. Arterioscler Thromb Vasc Biol 2018;38:e159-70. [Crossref] [PubMed]
- Liu X, Ma BD, Liu S, et al. Long noncoding RNA LINC00341 promotes the vascular smooth muscle cells proliferation and migration via miR-214/FOXO4 feedback loop. Am J Transl Res 2019;11:1835-42. [PubMed]
- Zhuang JB, Li T, Hu XM, et al. Circ_CHFR expedites cell growth, migration and inflammation in ox-LDL-treated human vascular smooth muscle cells via the miR-214-3p/Wnt3/β-catenin pathway. Eur Rev Med Pharmacol Sci 2020;24:3282-92. [PubMed]
- Lu QB, Wan MY, Wang PY, et al. Chicoric acid prevents PDGF-BB-induced VSMC dedifferentiation, proliferation and migration by suppressing ROS/NFκB/mTOR/P70S6K signaling cascade. Redox Biol 2018;14:656-68. [Crossref] [PubMed]
- García-Miguel M, Riquelme JA, Norambuena-Soto I, et al. Autophagy mediates tumor necrosis factor-α-induced phenotype switching in vascular smooth muscle A7r5 cell line. PLoS One 2018;13:e0197210. [Crossref] [PubMed]
- Jin X, Shi F, Chen Y, et al. Jet-Like Appearance in Angiography as a Predictive Image Marker for the Occlusion of Intracranial Atherosclerotic Stenosis. Front Neurol 2020;11:575567. [Crossref] [PubMed]
- van Hout GPJ, Bosch L. The Inflammasomes in Cardiovascular Disease. Exp Suppl 2018;108:9-40. [PubMed]
- Liu H, Chen H, Deng X, et al. Knockdown of TRIM28 inhibits PDGF-BB-induced vascular smooth muscle cell proliferation and migration. Chem Biol Interact 2019;311:108772. [Crossref] [PubMed]
- Dong X, Hu H, Fang Z, et al. CTRP6 inhibits PDGF-BB-induced vascular smooth muscle cell proliferation and migration. Biomed Pharmacother 2018;103:844-50. [Crossref] [PubMed]
- Bartoschek M, Pietras K. PDGF family function and prognostic value in tumor biology. Biochem Biophys Res Commun 2018;503:984-90. [Crossref] [PubMed]
- Chen S, Liu B, Kong D, et al. Atorvastatin calcium inhibits phenotypic modulation of PDGF-BB-induced VSMCs via down-regulation the Akt signaling pathway. PLoS One 2015;10:e0122577. [Crossref] [PubMed]
- Wang Z, Bhattacharya N, Weaver M, et al. Pim-1: a serine/threonine kinase with a role in cell survival, proliferation, differentiation and tumorigenesis. J Vet Sci 2001;2:167-79. [Crossref] [PubMed]
- Wang K, Deng X, Shen Z, et al. High glucose promotes vascular smooth muscle cell proliferation by upregulating proto-oncogene serine/threonine-protein kinase Pim-1 expression. Oncotarget 2017;8:88320-31. [Crossref] [PubMed]
- Willert M, Augstein A, Poitz DM, et al. Transcriptional regulation of Pim-1 kinase in vascular smooth muscle cells and its role for proliferation. Basic Res Cardiol 2010;105:267-77. [Crossref] [PubMed]
- Qattan A, Intabli H, Alkhayal W, et al. Robust expression of tumor suppressor miRNA's let-7 and miR-195 detected in plasma of Saudi female breast cancer patients. BMC Cancer 2017;17:799. [Crossref] [PubMed]
- Sayed AS, Xia K, Salma U, et al. Diagnosis, prognosis and therapeutic role of circulating miRNAs in cardiovascular diseases. Heart Lung Circ 2014;23:503-10. [Crossref] [PubMed]
- Zhou SS, Jin JP, Wang JQ, et al. miRNAS in cardiovascular diseases: potential biomarkers, therapeutic targets and challenges. Acta Pharmacol Sin 2018;39:1073-84. [Crossref] [PubMed]
- Liu Y, Fu W, Lu M, et al. Role of miRNAs in Epicardial Adipose Tissue in CAD Patients with T2DM. Biomed Res Int 2016;2016:1629236. [Crossref] [PubMed]
- Yang J, Li R, Zhao D, et al. Downregulation of microRNA-214 improves therapeutic potential of allogeneic bone marrow-derived mesenchymal stem cell by targeting PIM-1 in rats with acute liver failure. J Cell Biochem 2019;120:12887-903. [Crossref] [PubMed]
- Huang PS, Lin YH, Chi HC, et al. Thyroid hormone inhibits growth of hepatoma cells through induction of miR-214. Sci Rep 2017;7:14868. [Crossref] [PubMed]
(English Language Editor: J. Jones)


