
A novel 1-D densely connected feature selection convolutional neural network for heart sounds classification
Introduction
It is well known that early diagnosis of congenital heart disease (CHD) is directly related to the preservation of human health. Pathological/organic damage of the human cardiovascular system can be reflected in heart-related signals, for example in echocardiogram (ECG) and phonocardiogram (PCG) signals. Heart murmurs are important features associated to many of congenital heart disease, including regurgitation, stenosis of heart valves, left to right shunt lesions at the atrial, ventricular, or great arterial levels. Cardiac auscultation could differentiate normal heart sounds from abnormal pathological murmurs, therefore, it remains the most effective screening methods for congenital heart disease. Serious cardiac pathology may exist without symptoms so that many patients miss the optimal time for surgery when they are diagnosed. The main advantages for early recognizing a cardiac disease are that newborns will be seen and assessed earlier and in better clinical conditions. Then, based on the results of auscultation, the doctor decides whether to recommend further examinations, including echocardiogram, magnetic resonance, etc., to facilitate the diagnosis. In fact, clinically, it also needs to rely on the professional medical knowledge and skilled auscultation ability of doctors to determine heart health status or disease type of the patient. However, this usually requires a lot of time to train an advanced and skilled cardiovascular diagnostics specialist, which results in a severe shortage of cardiovascular specialists in lower-level hospitals and remote areas. Therefore, the need for objective and automatic auxiliary identification tools based on heart sound signals is particularly urgent.
Research on the automatic identification of cardiovascular disease types based on heart sound signals began more than 50 years ago (1,2). However, the field still faces many challenges. For traditional machine learning, the ability to effectively clean the original PCG signal data, remove complex and diverse noise, and extract identifiable features has become particularly important. In general, heart sound recognition methods involve two categories: recording level methods and fundamental heart sounds level (FHSS level) methods. For the FHSS level method, firstly, a segment of recorded heart sound is divided into a series of FHSS (a complete cardiac cycle or its estimation), with feature extraction then used to construct the classification and recognition model. Considering the non-steady characteristic of the PCG signal and the strong correlation between each cardiac cycle which constitutes a heart sound record, our study used a fixed-step overlapping sliding window to obtain the estimation fragment of FHSS, also called heart sound patches. Using this framework, our model is capable of realizing the data expansion and diversity, which results in the trained model having a stronger robustness and generalization ability. In addition, our model achieves end-to-end training, which makes the inference process of the model more efficient.
In recent years, deep learning techniques have shown impressive performance in many fields, including in audio, image, and video recognition, such as object detection (3,4), image segmentation (5), edge detection (6), and speech recognition (7). An increasing number of researchers are devoted to building networks with better expressiveness. At present, there is a general trend of focus gradually shifting from feature engineering to network topology engineering. A particularly noteworthy trend is that the proposed convolutional neural network (CNN) structure is getting deeper and deeper. Chen et al. (8) used modified frequency slice wavelet transform (MFSWT) to convert the one-dimensional cardiac cycle signal based on logistic regression hidden semi-Markov model (LR-HSMM) algorithm (9) into a two-dimensional time-frequency image and then combined two CNN models using sample entropy (SampEn) to select proper model for classifying normal and abnormal heart sounds. This proposed method achieved classification accuracy of 0.93 using 10-fold cross-validation on the PhysioNet/CinC Challenge 2016 dataset. However, this method relies on heart sound segmentation which means it is influenced by the accuracy of heart sound segmentation. It is a challenging task due to the complexity of PCG signals easily being contaminated by internal physiological noise and external noise (10). Furthermore, this method depends on the SampEn threshold which determine the selection of proper CNN model for classification. Therefore, inappropriate SampEn threshold would affect model selection and thus the prediction accuracy. The original LeNet5 (11) contained 5 layers, and VGGNet (12) has been upgraded to include 19 layers. In recent years, the proposed highway networks (13) and residual networks (14) have surpassed 100 layers, even reaching 1,000 layers. As the networks deepen, the performances of the models have been shown to significantly improve. However, further improvement of the network performance is directly hindered by the disappearance of gradients and degradation of network, which is due to the excessive depth of the network. The proposal of batch normalization (15), skip-layer connections between layers, has allowed, to a certain extent, solving of the problem of gradient disappearance and network degradation, making deeper network optimization possible. At the same time, a series of topologies for improving the flow of information have been proposed, such as deeply-supervised nets (16) and its variant (6), ResNet (14), inception-v4 (17) and so on, which allow further improvements of the performance of the deep CNN (DCNN).
There are an increasing number of researchers working on applying deep learning to the recognition of heart tone signals, along with how to build a more effective network for heart sound signal recognition. The CNNs are particularly good at fusing spatial and channel-wise information in order to extract effective features, but only for local receptive field fusion extraction, especially in the shallow part of the network. Recent studies have demonstrated that the performance of the network can be improved by embedding some modules that capture spatial information. A representative example is the inception architectures (17,18), which embed multi-scale processes in the modules to achieve competitive performance improvement. Based on recent developments of deep learning and the inherent characteristics of the PCG signals, the collected PCG signals have been shown to be susceptible to all kinds of noises from the external and internal environments; there are various types of heart murmurs, some of which are difficult even for experienced cardiovascular experts to judge by hearing only; the PCG signals are generally 10–20 s of heart sound recordings, which have obvious temporal and nonstationary properties. Thus, we proposed a dense feature selection convolution network (Dense-FSNet) which integrates Clique Blocks, SE block and SK-block performed on feature maps from convolution and inverse convolution operations, as a result of enhancing the information flow between low-level and high-level features, meanwhile extracting discriminative features from the fused multi-scale features for classification between normal and abnormal heart sounds.
Our proposed network architecture focuses on the following points: (I) the stacking of 4 cyclic densely connected Clique Blocks (19) ensure the maximum information flow between network layers. Unlike DenseNet, each layer is both an input and an output to the other layers, so that the information flow between low-level and high-level features is maximized in the module; (II) between 2 neighboring Clique Blocks, we also introduce the Squeeze-and-Excitation networks (SENet) (20) attention mechanism which is utilized for adaptively recalibrating channel-wise feature responses by modelling interdependencies between channels. During the process, by importing the attention mechanism, our model can strengthen the key features, whereas it weakens the irrelevant features, so as to alleviate the over-fitting drawback caused by over-parameterization of the deep network to a certain extent. Our SE Block can also be regarded as a kind of bypass connection, which can mitigate the effect of gradient disappearance; (III) through convolution and inverse convolution operations, the feature channel dimensions of shallow and deep modules are downscaled, and fusing features by element-wise-sum operation to build a multi-scale hyper-feature, which improves the classification capability of the network; (IV) we adopt an SK-block to model the fused multi-scale features and adaptively calibrate the final multi-scale features from a global perspective. Due to the network structure of cyclic dense connection and property of dual feature filtering, we refer to our approach as dense feature selection convolution network (Dense-FSNet).
Herein, we have performed a comprehensive evaluation of our proposed method with the PhysioNet computing in cardiology (CinC) 2016 challenge database (21). This the largest database of heart sound classifications accessible today. Compared with previous studies, our method was able to achieve state-of-the art levels which means it could achieve the highest accuracy when testing on the same database, and we present a detailed description and comparative analysis in the Methods and Results section. We present the following article in accordance with the STARD reporting checklist (available at https://dx.doi.org/10.21037/atm-21-4962).
Methods
Related work
The PCG analysis process generally involves several steps including heart sound pre-processing, heart cycle segmentation, feature extraction, and heart sound classification. Among these features, the accuracy of cardiac cycle segmentation is crucial for the classification performance. Cardiac cycle segmentation methods can be broadly divided into the following categories: methods based on envelope, methods based on feature extraction, and methods based on machine learning. Currently, the best method, logistic regression-hidden semi-Markov method (LR-HSMM) (9), is based on the hidden Markov model (HMM) theory. Although this method has achieved satisfactory results with publicly available datasets, it assumes that the cardiac cycle state conditions are independent, which does not fully reflect real-world conditions, along with requiring pre-processing with noise reduction to extract the heart sound features, which does not adapt well to the original heart sounds with more noise.
Traditionally, cardiac cycle segmentation and heart sound classification both involve feature extraction of the heart sound signal. Typically, three types of features are present in methods for heart sound feature extraction based on artificial design: time, frequency, and time-frequency domain-based features. Although these methods are easy to understand and compute, important information can be lost during the process. Furthermore, due to the non-stationary nature and diversity of heart sounds, it remains a challenge to extract more informative and discriminative feature representation from the original heart sound signals. The ultimate purpose of PCG analysis is to determine whether a heart sound has a pathological murmur, which is a heart sound classification problem. Methods used for heart sound classification include support vector machines (SVMs) (22,23), HMMs, k-nearest neighbors (KNNs) (22), neural networks (24-26), and other machine learning-based methods such as decision trees (27), Gaussian mixture models (GMMs) (28), and random forests (29). Whitaker et al. (23) extracted sparse coefficient matrices and time-domain features using SVM for heart sound type classification. Zhang et al. (30) used scaled spectrograms and partial least squares regression-based feature extraction in addition to support vector machine classifiers for heart disease detection. Hamidi et al. (22) used two different feature extraction methods, specifically Mel-frequency cepstrum coefficients (MFCCs) and fractal properties through pilling as features and used a KNN classifier based on Euclidean distance for classification. To further improve the classification performance, multi-classifier integration has also been introduced in several heart tone classification methods (24,31). Although the model-based integration approach may provide some improvement in classification accuracy, it also increases the computational complexity of the model, therefore making it more difficult to understand. Although traditional classifiers are simple to train and easy to deploy, they rely on handcrafted features that may not capture the most useful pattern of the PCGs.
In recent years, deep learning (DL) technology has rapidly developed. As one of the branches of DL, CNN technology has made remarkable achievements in many fields such as image, language, natural language processing (NLP), and so on by virtue of the strong feature representation power. In addition, the large open dataset of heart sounds PhysioNet/CinC 2016 (21) set the stage for the development of a DL-based classification method for heart sounds. Potes et al. (24) integrated AdaBoost’s variant classifier with CNN and subsequently won first place in the PhysioNet/CinC 2016 competition. Noman et al. proposed an ECNN method (26) that integrated one dimensional (1D) and 2D convolution. The 1D CNN can learn features directly from the original heart sound signal, while 2D CNN learns 2D time-frequency features based on MFCCs. Although these deep learning-based methods have been shown to achieve good classification performance, they lack a fine-grained architectural design and redundant model parameters, which are not sufficiently expressive to learn the complex patterns of PCGs. Xiao et al. (25) designed a 1D CNN architecture that was able to achieve good classification results without cardiac cycle segmentation. The CNN architecture exploits attentional mechanisms at both the spatial and channel levels to maintain a low number of parameters and obtain the distinguishing features. Inspired by CliqueNet (19), we have designed a new CNN architecture which was able to not only increase the CNN information flow, but also introduce a spatial attention mechanism. On the basis of integrating CNN multi-scale features, we were able to adaptively correct the weights of features at different scales, enabling the model to obtain a strong feature expression, and achieving excellent performance in PhysioNet/CinC 2016.
Data description
The heart sound signal public dataset PhysioNet/CinC 2016 (21) was used to construct and evaluate our model. The dataset contains 3,240 heart sound recordings, of which 2,939 are in the development set (2,425 and 514 positive abnormal heart sound recordings, respectively), and 301 heart sound recordings are in the independent test set. It should be noted that the dataset is composed of 6 different sources (which we denote as datasets a, b, c, d, e, and f). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Pre-processing
While deep CNNs have their own powerful expressive capabilities, as a result the networks require large amounts of data to drive the training of the models. Lack of data may result in a high risk of over-fitting. In addition, the multiple sources and significant imbalance of both the positive and negative sample distributions of the data set may also contribute to the training model focusing on categories with a large number of samples and “underestimating” categories with a small number of samples. Subsequently, this may affect the generalization ability of the model to test data. To solve this problem, we used an overlapping sliding intercept method (Figure 1) to amplify the data proportionally according to the data distribution.
Firstly, the PCG signals were down-sampled to 1,000 Hz, with the bands below 25 Hz and above 400 Hz respectively removed using a Butterworth band-pass filter. This resulted in the removal of both the low- and high-frequency noise in order to improve the signal-to-noise ratio. The development set was then processed as follows: analysis of the sample distribution of each sub-data set and determination that subset f was very different from the other 5 data sets and the sample size was very small, subset f was therefore treated as an outlier sample removal. Furthermore, in order to construct the final training set, each data subset was randomly sampled equally in another category according to the lesser amount of data (which was random 200 times). For each PCG signal, a series of heart sound patches were obtained by sliding the signal with a fixed window size and a fixed step size. This process was also used as the input for subsequent model training. As detailed in the Methods and Experiments section, a sliding window with a window size of 800 and a step size of 200 was used to obtain our patch data set. By using this approach, we were able to greatly increase both the sample size and diversity of the samples in order to make the model more robust to acquisition.
Proposed CNN
As shown in Figure 2, the backbone network of our proposed Dense-FSNet consists of a stack of 4 cyclically densely connected Clique Blocks. Behind the first 3 Clique Blocks, channel-wise feature adaptive activation processing via SE block was performed, with the model ending proposed. Subsequently, the multi-scale attention mechanism adaptively activates and then fuses features from different scales of the model to form the final classification features.
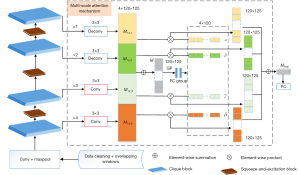
Clique blocks
Inspired by CliqueNet (19), we successfully built the Clique Blocks. This algorithm uses a bi-directional circular dense network module, an architecture inspired by the long-short term memory (LSTM) and attention mechanism in which each layer is both an input and an output of the other layers. By using this architecture, our model is better able to focus on features relevant to the training task by multiplexing the feature maps of the convolutional output. In addition, this forward and backward connection between the convolutional layers can greatly enhance the information flow in the deep network to take full advantage of the spatiotemporal information contained in the data. Each module of the Clique Block can be divided into two phases. The first stage is propagated similar to DenseNet, which in this context can be viewed as the initialization process. In this stage, the output of the front layer takes the input of the back layer, and the data flows from the front layer to the back layer according to the diagram of the upper sub-figure. In the second stage, the input of each convolutional operation includes not only the output features of all previous layers, but also those of subsequent layers. The second stage of the convolutional feedback structure uses higher level visual information to refine the features of the previous stage in order to achieve spatial focus. In this stage, the output data flow of each updated layer flows again to the last layer according to the diagram of the lower sub-figure, and the output of the last layer takes the output of the whole Clique Block.
Squeeze-and-excitation block
It is known that the core of DCNN is based on convolutional operations, which are essentially the fusion of spatial and channel dimensional information of local receptive fields. Our proposed Dense FSNet backbone module, Clique Block, greatly enhances feature multiplexing and information flow in the network through cyclic dense connections. However, it is also prone to over-parameterization, which may lead to model overfitting. While, the SENet proposed by Hu et al. (20) could improve the representational capacity of a network by enabling it to preform dynamic channel wise feature recalibration. Firstly, the module performs a GAP (global average pooling) operation on the input feature map X, thus aggregating the global spatial information for the channel representation, which can also be considered as the statistics of each channel feature. In order to simplify the operation, the bottleneck structure was constructed with two FC layers. The sigmoid activation function is used to obtain the activation weights of each channel feature. The final output of this module is the result of the Scale operation on the input and activation weights of the module, which enables adaptive correction of each channel feature, and can also be understood as channel-wise feature filtering.
Multi-scale attention mechanism
It is widely accepted that the features extracted from CNN networks become increasingly abstract as the network deepens, and it becomes progressively more important to combine the low-level visual features with the high-level semantic features to build a more effective network framework for different business scenarios. Our proposed Dense-FSNet goes through four cyclic densely connected modules that output 1/3, 1/6, 1/12, and 1/24 features relative to the input, compared to other studies that construct multi-scale hyper-features via element-wise sum or concatenate operations. The Multi-Scale Attention Mechanism module (Figure 2) is able to not only construct hyper-features, but also adaptively adjust the proportional weights of various scale features in the hyper-feature according to the different stimuli received by the neuron. In turn, these can be used to globally analyze the Multi-Scale Attention Mechanism. The 4 feature groups of different scales exported by Base-Net are unified to 120×125 dimensions by a convolutional layer with a kernel size of 3, corresponding to respectively, noting the hyper-feature as M:
The hyper-feature then goes through a global average pooling layer (GAP) in order to generate the statistics v of the feature, where :
The bottleneck structure is constructed through the FC layer, with the number of channels first halved to C/2 and then boosted to C. Finally, the adaptive activation vectors [α β γ δ] for each multi-scale feature are generated through the softmax layer. The activation vector is then multiplied with the set of features of each scale to obtain a new adaptively corrected hyper-feature i.e., Ffinal for the final PCG signal classification identification.
Statistical analysis
The experimental design used here was divided into two main parts in order to: (I) compare the training and testing performance of our proposed model algorithm and the current algorithm with better results (1-3) on the same dataset, and (II) conduct a screening comparison experiment on the main hyper parameters of our proposed model framework. Through systematic comparative analysis, we found that our proposed Dense-FSNet achieved a significant improvement in the recognition of abnormal heart sounds when compared to other algorithms. In order to verify the robustness of the model, a 10-fold cross validation experiment was also conducted. Finally, a 1D Grad-Cam was used to visualize the activation of the predictions to help us understand the model and enhance the interpretability of the results, as shown in the Results section.
The model training process is patch-based, so a patch-bag for testing was also generated, and then voted to determine the final category (the threshold is set to 0.5, i.e., if more than half of the patches are abnormal, the segment is considered abnormal and vice versa). The effect of the model was assessed using the accuracy, specificity, sensitivity, and area under the curve (AUC) values, as follows:
Where true positive (TP): positive samples predicted to be positive; false positive (FP): negative samples that are predicted to be positive; false negative (FN): positive samples that are predicted to be negative; true negative (TN): negative samples that are predicted to be negative.
Results
Evaluation on the test set
The heart sound public dataset PhysioNet/CinC 2016 independent test set contains a total of 301 heart sound recordings, of which 151 are abnormal heart sounds and 150 are normal heart sounds. In order to better compare and analyze the performance of our proposed Dense-FSNet in heart sound signal recognition, we conducted systematic experiments to compare it with several current cutting-edge algorithms, including the fusion model of AdaBoost and CNN proposed by PhysioNetCinC 2016 challenge winner Cristhian Potes (24). These models achieved accuracy, specificity, and sensitivity of 0.827, 0.69, and 0.97 respectively on an independent test set. Bin Xiao (25) proposed a 1-D convolutional model for end-to-end training prediction, which achieved accuracy, specificity, and sensitivity of 0.814, 0.74, and 0.89 respectively. Similarly, Noman et al. (26) proposed the ECNN model based on the eigen gram of heart sound signals, which achieved accuracy, sensitivity, and specificity of 0.811, 0.810, and 0.810, respectively. When compared to other algorithms, our proposed Dense-FSNet model achieved an accuracy, specificity and sensitivity of 0.867, 0.94 and 0.79 respectively, which is 4.01% higher than that of Cristhian Potes (24) and 5.61% and 5.31% higher than that of ECNN (26) and Bin Xiao (25), respectively. The accuracy for each algorithm is presented in Table 1. In conclusion, our model has significant performance advantages.
Table 1
In order to compare and analyze more intuitively, we also analyzed the changes in the accuracy of 1D-convolution, ECNN, and Dense-FSNet with increasing number of iterations during the training process. According to Figure 3, the model of 1D-covnlution and ECNN performance stabilized after 20 epochs of iterations, whereas Dense-FSNet continued to improve by 20 epochs before converging and achieving the best results.
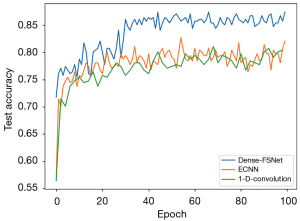
The results of receiver operating characteristic (ROC) statistical analysis for Dense-FSNet, ECNN, and 1D-convolution are shown in Figure 4. The AUC value of our proposed algorithm is the highest, reaching 0.93, whereas the other 2 algorithms are 0.88 and 0.91, respectively.
Super-parameter selection
The algorithmic framework proposed in this study, Dense-FSNet, has several key hyperparameters that need to be adjusted. One of key hyperparameters is the window size and step size when performing sliding window to get heart sound patches. In this paper, we use a fixed size step size [200], and the grid search method was used to select the window size. We found that when the window size is 800, the performance of the model is basically optimal, and further increasing the window size does not improve or even leads to performance degradation. Further analysis revealed that since our data down-sampled to 1,000 Hz, 800 sample points covered one cardiac cycle of most PCG signals, which also supported the rationality of the window size of 800. We also compared and analyzed whether adding the Multi-Scale Attention Mechanism module to our proposed Base-Net had an impact on the predictive performance of the model. The results showed that adding the Multi-Scale Attention Mechanism module was able to significantly improve the performance of the model. It is noteworthy that due to the multi-source nature of the PhysioNet/CinC 2016 dataset and the extreme imbalance of positive and negative samples, it becomes crucial to make a reasonable data balance during the model training process. Detailed comparison results can be found in Table 2.
Table 2
| Window size | Step size | Multi-scale attention | Data balance | Acc. | Sen. | Spe. |
|---|---|---|---|---|---|---|
| 600 | 200 | Y | Y | 0.8432 | 0.93 | 0.75 |
| 1,000 | 200 | Y | Y | 0.8638 | 0.95 | 0.78 |
| 1,200 | 200 | Y | Y | 0.8505 | 0.93 | 0.77 |
| 800 | 200 | Y | Y | 0.8671 | 0.94 | 0.79 |
| 800 | 200 | Y | N | 0.8272 | 0.80 | 0.85 |
| 800 | 200 | N | Y | 0.8472 | 0.92 | 0.77 |
Dense-FSNet, Dense Feature Selection Convolution Network framework.
Cross validation result
To better validate the robustness of the comparison algorithms, we performed a 10-fold cross validation analysis of 1D-convolution (25), ECNN (26), and our proposed Dense-FSNet on the PhysioNet/CinC 2016 dataset. The development set was divided into 10 equal parts, each time using 9 training and 1 testing, and finally averaged. From the experimental results, as Table 3 showed, the accuracy, sensitivity, and specificity of 1D-convolution cross validation were 0.93, 0.86, and 0.95, respectively; the accuracy, sensitivity, and specificity of ECNN cross validation were 0.9191, 0.933, and 0.87, respectively; the Dense-FSNet proposed in this paper was still able to achieve the highest accuracy, sensitivity, and specificity in the cross-validation session. The accuracy, sensitivity and specificity were determined to be 0.9509, 0.885, and 0.97, respectively, which once again supports the superiority of our proposed algorithm in the heart tone signal recognition project.
Table 3
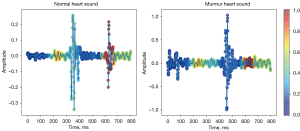
Heart sound recognition heatmap
In order to better understand which heart sound segments are used to obtain the classification results when a particular model makes a decision, we elected to use Grad-CAM (32) to visualize the heart sound regions of interest to the model. Our results showed that when the model predicted normal heart sound, it tended to pay attention to the information of S1 and S2 regions of heart sound, whereas when the model predicted abnormal heart sound, it activated different regions according to the location of murmur (as shown in Figure 5). It should be noted that due to the complexity of the heart sounds, the regions activated by the model are difficult to understand for some samples, despite the general trend described above. Nonetheless, the use of Grad-CAM to visualize the regions of heart sounds that the model relies on making decision provides some medical interpretability to the model, which can be useful for subsequent model improvement.
Discussion
In this study, we constructed a new deep CNN for the detection and identification of abnormal heart sounds that was capable of obtaining results significantly better than current models. The framework constructed, Dense-FSNet, is able to enhance the information flow of the entire network through employment of the circular dense connection module Clique Block, which serves to fully improve the feature utilization efficiency and reduce feature redundancy. Subsequently, channel-wise feature adaptive correction was used to strengthen critical features and weaken irrelevant or minor features so as to prevent over-parameterization of the model and to filter local features. At the end of the network, we also used the Multi-Scale Attention Mechanism in order to not only fuse the features from different scales of the network to obtain the Hyper Feature, but also use the Attention Mechanism to adaptively correct the weights of different scales to perform global feature filtering for the final heart sound recognition. Experimental evaluations on the dataset PhysioNet/CinC 2016 showed that our proposed algorithm achieved better performance than the current state-of-the-art method, which demonstrates the potential and significance of building deep networks suitable for heart tone signal recognition.
Dense-FSNet supplied 95% effectiveness for discriminating heart sound data as innocent and pathological murmurs, which means that 95 patients out of 100 were correctly diagnosed. When an innocent murmur is detected, it does not represent current or future illness, unless it is pathological. But a physician often needs to order an echocardiogram for reassurance, whereas primary health care institutions or low-level hospitals are often not equipped with echocardiography equipment. This analysis of heart sounds provides information that can be very useful for a physician when deciding whether or not to release the patient or to send him/her for an echocardiogram. The limitation is that when pathological murmurs are detected by this method, it still needs to be combined with clinical history, physical examination and imaging examination to facilitate the diagnosis. Meanwhile, electrocardiograms are not routinely used to screen for or diagnose congenital heart disease unless the child has symptoms or disease associated with arrhythmia. We acknowledge that our framework is not only limited to the classification of one-dimensional audio data like PCG, but may also be extended to other artificial intelligence (AI) domains, including image recognition, target detection, and speech recognition. Based on these results, we propose that additional research will combine LSTM and the idea of multi-model fusion to further investigate the application of deep learning in heart sound signal recognition, and also extend our proposed network model to other research directions. This combined innovation model may bring higher accuracy and sensitivity in the application of heart sound recognition.
Acknowledgments
We would like to thank Qin Gao and Zhaoxi Wang from Xinhua Harvard International Healthcare Innovation Collaboration Initiatives for manuscript discussion and review.
Funding: The project was funded by Research project of Shanghai Translational Medicine Collaborative Innovation Center (TM202007) and Clinical innovation project of Xinhua Hospital (19XHCR14D).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-4962
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-4962). XW, XL, YZ are from Ewell Technology Co., Ltd. JW is from Xunyin Intelligent Technology (Shanghai) Co., Ltd. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gerbarg DS, Taranta A, Spagnuolo M, et al. Computer analysis of phonocardiograms. Prog Cardiovasc Dis 1963;5:393-405. [Crossref]
- Liang H, Nartimo I. A feature extraction algorithm based on wavelet packet decomposition for heart sound signals. IEEE International Symposium on Time-Frequency and Time-Scale Analysis 1998:93-6.
- Girshick R, Donahue J, Darrell T, et al. Region-Based Convolutional Networks for Accurate Object Detection and Segmentation. IEEE Trans Pattern Anal Mach Intell 2016;38:142-58. [Crossref] [PubMed]
- Girshick R, Donahue J, Darrell T, et al. Rich Feature Hierarchies for Accurate Object Detection and Semantic Segmentation. IEEE Conference on Computer Vision and Pattern Recognition 2014:580-7.
- Shelhamer E, Long J, Darrell T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans Pattern Anal Mach Intell 2017;39:640-51. [Crossref] [PubMed]
- Xie S, Tu Z. Holistically-Nested Edge Detection. IEEE International Conference on Computer Vision (ICCV) 2015:1395-403.
- Chen X, Liu X, Wang Y, et al. Efficient Training and Evaluation of Recurrent Neural Network Language Models for Automatic Speech Recognition. IEEE Transactions on Audio Speech and Language Processing 2016;24:2146-57. [Crossref]
- Chen Y, Wei S, Zhang Y. Classification of heart sounds based on the combination of the modified frequency wavelet transform and convolutional neural network. Med Biol Eng Comput 2020;58:2039-47. [Crossref] [PubMed]
- Springer DB, Tarassenko L, Clifford GD. Logistic Regression-HSMM-Based Heart Sound Segmentation. IEEE Trans Biomed Eng 2016;63:822-32. [PubMed]
- Leal A, Nunes D, Couceiro R, et al. Noise detection in phonocardiograms by exploring similarities in spectral features. Biomed Signal Proces 2018;44:154-67. [Crossref]
- Lecun Y, Bottou L, Bengio Y, et al. Gradient-based learning applied to document recognition. P IEEE 1998;86:2278-324. [Crossref]
- Russakovsky O, Deng J, Su H, et al. ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vision 2015;115:211-52. [Crossref]
- Sirvastava R, Greff K, Schmidhuber J. Training Very Deep Networks. Advances in Neural Information Processing Systems 2015;2377-85.
- He K, Zhang X, Ren S, et al. Deep Residual Learning for Image Recognition, Computer Vision and Pattern Recognition. IEEE Conference on Computer Vision and Pattern Recognition 2016:770-8.
- Ioffe S, Szeged C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. Proceedings of the 32nd International Conference on International Conference on Machine Learning 2015;1:448-56.
- Lee C, Xie S, Gallagher P, et al. Deeply-Supervised Nets. J Mach Learn Res 2015;38:562-70.
- Szegedy C, Ioffe S, Vanhoucke V, et al. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. The Thirty-First AAAI Conference on Artificial intelligence 2017:4278-84.
- Szegedy C, Vanhoucke V, Ioffe, S et al. Rethinking the Inception Architecture for Computer Vision. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 2016:2818-26.
- Yang Y, Zhong Z, Shen T, et al. Convolutional Neural Networks with Alternately Updated Clique. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition 2018:413-22.
- Hu J, Shen L, Albanie S, et al. Squeeze-and-Excitation Networks. IEEE Trans Pattern Anal Mach Intell 2020;42:2011-23. [Crossref] [PubMed]
- Liu C, Springer D, Li Q, et al. An open access database for the evaluation of heart sound algorithms. Physiol Meas 2016;37:2181-213. [Crossref] [PubMed]
- Hamidi M, Ghassemian H, Imani M. Classification of heart sound signal using curve fitting and fractal dimension. Biomed Signal Proces 2018;39:351-9. [Crossref]
- Whitaker BM, Suresha PB, Liu C, et al. Combining sparse coding and time-domain features for heart sound classification. Physiol Meas 2017;38:1701-13. [Crossref] [PubMed]
- Potes C, Parvaneh S, Rahman A, et al. Ensemble of feature-based and deep learning-based classifiers for detection of abnormal heart sounds. Computing in Cardiology Conference 2016:621-4.
- Xiao B, Xu Y, Bi X, et al. Heart sounds classification using a novel 1-D convolutional neural network with extremely low parameter consumption. Neurocomputing 2018;392:153-9. [Crossref]
- Noman F, Ting C, Salleh S, et al. Short-segment Heart Sound Classification Using an Ensemble of Deep Convolutional Neural Networks. IEEE International Conference on Acoustics Speech and Signal Processing-Proceedings 2019:1318-22.
- Varghees V, Ramachandran K. Effective Heart Sound Segmentation and Murmur Classification Using Empirical Wavelet Transform and Instantaneous Phase for Electronic Stethoscope. IEEE Sens J 2017;17:3861-72. [Crossref]
- Mannini A, Sabatini AM. Machine learning methods for classifying human physical activity from on-body accelerometers. Sensors (Basel) 2010;10:1154-75. [Crossref] [PubMed]
- Atallah L, Lo B, King R, et al. Sensor positioning for activity recognition using wearable accelerometers. IEEE Trans Biomed Circuits Syst 2011;5:320-9. [Crossref] [PubMed]
- Zhang W, Han J, Deng S. Heart sound classification based on scaled spectrogram and partial least squares regression. Biomed Signal Proces 2017;32:20-8. [Crossref]
- Nabhan Homsi M, Warrick P. Ensemble methods with outliers for phonocardiogram classification. Physiol Meas 2017;38:1631-44. [Crossref] [PubMed]
- Selvaraju R, Cogswell M, Das A, et al. Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization. Int J Comput Vis 2020;128:336-59. [Crossref]
(English Language Editor: J. Jones)




