
Identification of solute carrier family genes related to the prognosis and tumor-infiltrating immune cells of pancreatic ductal adenocarcinoma
Introduction
Pancreatic cancer (PC) is the fourth and the sixth leading cause of cancer-related death in the US and China, respectively (1,2). In 2018, there were approximately 458 million new cases and 432 million deaths due to PC (3). The above statistics indicate that PC is a highly malignant neoplasm with a similar morbidity and mortality; and the 5-year survival rate remains low, at 6% (4). Pancreatic ductal adenocarcinoma (PDAC) is the predominant pathological type of PC (5). Patients with PDAC are typically first diagnosed at an advanced stage, and thus, have surpassed the opportunity for surgical treatment. This is because patients do not exhibit classic symptoms at an early stage and there is an absence of powerful detective biomarkers (6). Some patients are lucky and undergo radical resection; however, the 5-year survival rate is only as high as 25% (7). Thus, new biomarkers are required to improve outcomes.
The solute carrier (SLC) gene superfamily is the second largest family of membrane proteins after G protein-coupled receptors, including over 400 proteins in 65 subfamilies based on sequence similarities (8). Amino acids, sugars, fatty acids, inorganic ions, essential metals, and drugs are transported over the cell membrane by the SLCs, which function as passive transporters, ion transporters, and exchangers (9). Since SLCs are responsible for transporting essential substances throughout the human body, SLC mutations have been linked to human genetic disorders and the level of SLC expression is changed in a variety of tumors, including congenital chloride diarrhea, glucose galactose malabsorption, and familial renal glucosuria (10-12). A study showed that SLC5A8 could suppress colon cancer and was silenced by methylation (13). In a mouse model, reducing polyamine was effective for prolonging prognosis and preventing tumor progression, and a knockdown of SLC3A2 in neuroblastoma cells reduced the uptake of polyamine (14). Moreover, SLCs have been extensively studied in pharmacokinetics, chemotherapy resistance, and as therapeutic targets (15). Nucleoside transporters encoded by the SLC28 and SLC29 subfamilies have been shown to mediate the uptake of gemcitabine and 5-fluorouracil. In addition, some of the SLC22A subfamily is related to the transport of platinum compounds (16,17). Mutations of SLC affect the synthesis of transporters, finally resulting in an insufficient intake of chemotherapeutic drugs which will inevitably lead to decreased sensitivity to chemotherapy. Although a previous study found that SLC2A1 may be a prognostic biomarker for PDAC, the SLC family played an important role in biological processes such as metabolism, and the comprehensive analysis of the SLC family in PDAC was still unknown (18). As well as, the potential mechanism of most members of the SLC family in PDAC and the relationship between genes and the prognosis of PDAC remains unclear.
The tumor microenvironment (TME) consists of multiple cell types (e.g., endothelial cells, immune cells, and so on) and extracellular components. Tumor cells can alter the TME through immune escape and immunosuppression; thus, the TME plays a critical role in the occurrence, progression, and treatment of tumors (19,20). In addition, immune cells are an essential cell type involved in TME and targeting immune cells is a promising therapy in PC (21). The effect of PD1 or PD-L1 checkpoint inhibitor immunotherapy has improved the clinical outcomes in a variety of tumors, including advanced melanoma, Hodgkin’s lymphoma, and advanced gastric or gastro-esophageal junction cancer (22-24). However, targeting immune checkpoints has not benefited all patients in a variety of tumors, including PDAC (25). Researching the differences in tumor-infiltrating immune cells (TIICs), the relationship with molecular expression and interactive features in individual tumors, and therefore, the identification of new immunotherapeutic targets, is critical for improving patient prognosis. Studies shown that the tumor microenvironment of PDAC was highly heterogeneous, and PD-1+ cells, and Foxp3+ T cells could evaluate the prognosis of patients with PDAC after surgery (26,27). A study shown that SLC1A5, SLC7A5, SLC3A2 were associated with the expression of PD1 and PD-L1, and might be associated with subtypes of immune cell infiltration. Therefore, we speculated that the SLC family could play a role in the TME of PDAC, especially TIICs (28).
In this study, we collected data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases, using bioinformatics tools to explore the relationship between SLC family genes and prognosis of PDAC patients, the underlying molecular mechanisms, and the density of numerous TIICs associated with clinicopathological characteristics. These results are promising for providing a new perspective on the TME and identifying potential biomarkers to achieve better clinical outcomes of PDAC.
We present the following article in accordance with the TRIPOD reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6341/rc).
Methods
Data collection
We downloaded RNA-Seq expression profiles and clinical data from the publicly available TCGA (https://cancergenome.nih.gov/) and the University of California, Santa Cruz Xena (UCSC Xena: https://xena.ucsc.edu/) databases, respectively (29). The raw expression profiles were normalized using DESeq package in R (29). The following inclusion criteria were previously published by our research team: (I) complete survival data available; (II) histology type was PDAC; (III) pathologic stage I or II; and (IV) patients underwent pancreaticoduodenectomy. Any PDAC patients with pathologic stage III or IV disease who underwent other types of surgery were excluded (30). To investigate whether there are differences in gene expression in the peripheral blood (PB) and peripheral blood mononuclear cells (PBMCs), GSE49641 and GSE74629 were downloaded from the GEO database. If there were multiple expression values for the same gene name, the average value and expression profiles were normalized using the limma package in R (https://bioconductor.org/packages/release/bioc/html/limma.html). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Survival analysis
A Kaplan-Meier analysis was used to assess prognosis-related clinical factors. Patients were divided into low- and high-expression groups according to the median gene expression value. We used Kaplan-Meier analysis to preliminarily screen for the genes related to prognosis. Next, a least absolute shrinkage and selection operator (LASSO) algorithm was used to select the strongest prognostic-related genes. Prognostic-related genes were finally defined by a Cox proportional risk regression model that was adjusted by prognostic-related clinical factors.
Bioinformatics analysis
The Gene Expression Profiling Interactive Analysis (GEPIA; https://gepia.cancer-pku.cn/index.html) website was used to evaluate differences in gene expression in both the tumor and non-tumor tissues (31). The protein-protein interaction (PPI) network, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was obtained from the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; https://string-db.org/, version 11) (32). The results of the LASSO analysis of SLCs were entered into the website, and further obtained after setting Homo sapiens and an interaction score >0.150. The KEGG results were visualized using R. We then used GeneMANIA (https://genemania.org/) to acquire details of gene-gene interactions (33). A Pearson’s correlation coefficient between the SLC genes was calculated, and the corrplot package in R (https://cran.r-project.org/web/packages/corrplot/index.html) was used to visualize the results. The differences in SLC expression between PDAC and healthy controls in PB and PBMCs were analyzed and plotted using the ggplot2 package in R (https://cran.r-project.org/web/packages/ggplot2/index.html).
Joint-effect analysis and prognostic signature construction
To increase the applicability of the results, we used the prognostic-related genes to establish a joint-effect analysis with a Kaplan-Meier analysis. Nomograms were constructed using clinical variables and prognostic-related genes were used to predict the overall survival (OS) for PDAC using the rms package (https://cran.r-project.org/web/packages/rms/index.html). A prognostic risk model was constructed using prognostic-related genes. SLC19A3, SLC25A39, and SLC39A11 were included in nomogram because only they satisfied prognosis-related genes and differential expression in both the tumor and normal tissues. Regression coefficients were obtained from the results of the multivariate Cox proportional hazards regression analysis. The calculation formula for the risk score was as follows (30):
Risk score = expression of gene1 × β1 + expression of gene2 × β2+… expression of Genen × βn.
The participants were divided into high- and low-risk groups based on their risk score. Receiver operating characteristic (ROC) curves were constructed to assess the accuracy of the prognostic signature using prognostic-related genes and risk score. A forest plot was constructed to assess whether the risk model was combined with other clinical factors, which has an impact on the prognosis.
CIBERSORT estimation
To investigate the TIIC landscape in the PDAC tissue between the high- and low-risk groups, the Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) algorithm (https://cibersort.stanford.edu) was used to calculate the absolute proportions of 22 types of TIICs (34). CIBERSORT, which contained 547 genes, could accurately differentiate 22 individual immune cell types from the tumor is a deconvolution algorithm and is the most widely used method to date (35). A normalized PDAC dataset was input as a mixture file, 22 immune cell types (LM22) were set as the signature gene file, and an analysis was performed at 1,000 permutations. The samples which resulted in P<0.05 were considered statistically significant. We used the corrplot package in R to visualize the Pearson’s correlation coefficient between different immune cells. A bar graph was constructed using barplot packages to demonstrate a landscape of TIICs for statistically significant cases. The differences in TIICs between high- and low-risk groups were shown in a violin plot.
Gene set enrichment analysis (GSEA)
GSEA is an algorithm that can use known gene expression to calculate the potential mechanisms of differential gene expression, affecting the prognosis of patients (36). High- and low-group files determined by prognostic-related genes and genome-wide expression profile were uploaded to the GSEA. We included C2 and C5 of the Molecular Signatures Database (MSigDB) in the enrichment analysis (37). Meeting both the false discovery rate (FDR) <0.25 and P<0.05 was considered statistically significant evidence (38).
Statistical analysis
The statistical analyses were conducted using SPSS software version 22.0 (IBM Corp., Armonk, NY, USA) and R-project version 3.6.3 (https://cran.r-project.org/bin/windows/base/old/3.6.3/). The Kaplans://cr method was used to calculate the median survival time and log-rank P value. Hazard ratios (HRs) and 95% confidence intervals (CI) were calculated using the univariate and multivariate Cox proportional hazards regression model. A P value <0.05 was considered statistically significant.
Results
Data collection
A total of 183 pancreatic adenocarcinoma (PAAD) cases were downloaded, and after applying the inclusion and exclusion criteria, 112 PDAC cases were finally included in the follow-up research in TCGA. In total, 36 and 50 PDAC and non-tumor patients, respectively, were included from GSE49641 and GSE74629 in the subsequent analyses.
Survival analysis
Prognosis-related clinical variables are shown in Table S1, and the histologic grade, targeted molecular therapy, radiation therapy, and residual resection were shown to significantly affect the patient’s prognosis. The results of the prognostic differences from the low- and high-expression groups of each SLC are presented in Table S2. In addition, 32 SLC genes were significantly associated with the OS. The genes SLC19A3, SLC22A23, SLC22A4, SLC25A11, SLC25A39, SLC26A10, SLC35B4, SLC35E2, SLC39A11, SLC46A2, SLC47A1, and SLC52A1 were the results of the LASSO analysis (Figure 1A,1B and Figure S1). After calculation using a Cox proportional risk regression model, SLC19A3 (adjusted P=0.007), SLC22A23 (adjusted P=0.035), SLC25A39 (adjusted P=0.027), SLC39A11 (adjusted P=0.043), and SLC47A1 (adjusted P=0.024) were significantly correlated with the prognosis (Table 1).
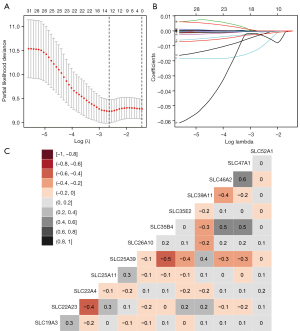
Table 1
| Gene expression | Events/total (n=112) | MST (days) | Crude HR (95% CI) | Crude P value | Adjusted HR (95% CI) | Adjusted P valuea |
|---|---|---|---|---|---|---|
| SLC19A3 | ||||||
| Low | 39/56 | 449 | 1 | 1 | ||
| High | 30/56 | 756 | 0.460 (0.276–0.766) | 0.003 | 0.447 (0.250–0.800) | 0.007 |
| SLC22A23 | ||||||
| Low | 43/56 | 497 | 1 | 1 | ||
| High | 26/56 | 756 | 0.447 (0.272–0.732) | 0.001 | 0.526 (0.289–0.956) | 0.035 |
| SLC22A4 | ||||||
| Low | 38/56 | 506 | 1 | 1 | ||
| High | 31/56 | 732 | 0.581 (0.357–0.945) | 0.029 | 0.676 (0.397–1.151) | 0.149 |
| SLC25A11 | ||||||
| Low | 38/56 | 554 | 1 | 1 | ||
| High | 31/56 | 681 | 0.598 (0.368–0.971) | 0.038 | 0.647 (0.382–1.096) | 0.105 |
| SLC25A39 | ||||||
| Low | 30/56 | 748 | 1 | 1 | ||
| High | 39/56 | 503 | 1.880 (1.153–3.065) | 0.011 | 1.852 (1.071–3.202) | 0.027 |
| SLC26A10 | ||||||
| Low | 43/56 | 549 | 1 | 1 | ||
| High | 26/56 | 684 | 0.613 (0.375–1.003) | 0.051 | 0.931 (0.528–1.641) | 0.804 |
| SLC35B4 | ||||||
| Low | 37/56 | 504 | 1 | 1 | ||
| High | 32/56 | 714 | 0.582 (0.356–0.953) | 0.031 | 0.707 (0.402–1.244) | 0.229 |
| SLC35E2 | ||||||
| Low | 42/56 | 534 | 1 | 1 | ||
| High | 27/56 | 754 | 0.584 (0.356–0.955) | 0.032 | 0.674 (0.401–1.133) | 0.137 |
| SLC39A11 | ||||||
| Low | 32/56 | 705 | 1 | 1 | ||
| High | 37/56 | 538 | 1.699 (1.046–2.761) | 0.032 | 1.749 (1.017–3.009) | 0.043 |
| SLC46A2 | ||||||
| Low | 37/56 | 524 | 1 | 1 | ||
| High | 32/56 | 727 | 0.549 (0.336–0.895) | 0.016 | 0.783 (0.454–1.350) | 0.378 |
| SLC47A1 | ||||||
| Low | 39/56 | 459 | 1 | 1 | ||
| High | 30/56 | 769 | 0.409 (0.249–0.670) | <0.001 | 0.504 (0.278–0.913) | 0.024 |
| SLC52A1 | ||||||
| Low | 40/56 | 544 | 1 | 1 | ||
| High | 29/56 | 688 | 0.611 (0.376–0.993) | 0.047 | 0.774 (0.430–1.289) | 0.292 |
Italic P values indicate statistically significant. a, adjusted for histologic grade, radiation therapy, radical resection, and targeted molecular therapy. SLC, solute carrier; MST, median survival time; OS, overall survival; PDAC, pancreatic ductal adenocarcinoma; HR, hazard ratio; CI, confidence interval.
Bioinformatics analysis
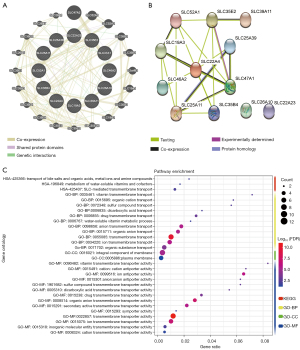
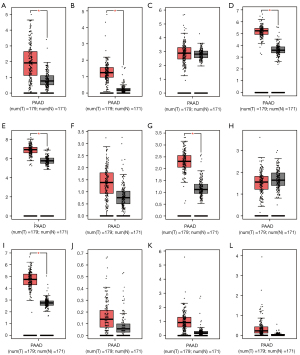
The Pearson’s correlation coefficient from the 12 genes selected from LASSO analysis are presented in Figure 1C. The genes SLC47A1 and SLC46A2, SLC35B4 and SLC47A1, and SLC46A2 were moderately positive in relation to each other (coefficient >0.5), whereas SLC35B4 was moderately negative in relation to SLC25A39 (coefficient <−0.5). The gene-gene interactions displayed in Figure 2A show a strong co-expression relationship between these genes. The PPI is presented in Figure 2B and shows interactions in expression, experiment, text-mining, and co-expression. The GO and KEGG pathway enrichment analyses were mainly enriched in acids, inorganic ions, and essential metal transmembrane transporter activity (Figure 2C). The distribution in SLC expression between PAAD tumor and normal tissues from GEPIA suggested that the expression of SLC39A11, SLC25A39, SLC22A4, SLC35B4, SLC25A11, and SLC19A3 were significantly higher in the tumor tissue (Figure 3). In PB, the level of SLC22A4, SLC25A11, and SLC46A2 expression in PDAC patients was higher than that in the control samples (Figure S2). In PBMCs, only SLC35E2 exhibited significantly higher expression in PDAC patients (Figure S3).
Joint-effect analysis and prognostic signature construction
According to the above study, we found that only SLC19A3, SLC25A39, and SLC39A11 satisfied prognosis-related genes and differential expression in both the tumor and normal tissues. Thus, a joint-effect analysis and prognostic signature were comprised of SLC19A3, SLC25A39, and SLC39A11. In the joint-effect analysis, compared with Group A, Group III and Group 4 had the best prognosis in their large group (Table 2). We constructed a nomogram using clinical factors and the 3 SLC genes mentioned above which can be used to predict the 1-, 2-, and 3-year OS of PDAC patients (Figure 4A). The forest plot indicated that patients in the low-risk group had a significantly better prognosis than the high-risk group under the following conditions: young patients (young patients condit alcohol history, high histologic grade (G3 + G4), no radical resection, no radiation therapy, and those who received targeted macular therapy (Figure 4B).
Table 2
| Group | SLC19A3 | SLC25A39 | SLC39A11 | Patients | Number of events | MST | Crude HR (95% CI) | Crude P value | Adjusted HR (95% CI) | Adjusted P valuea |
|---|---|---|---|---|---|---|---|---|---|---|
| A | Low | High | – | 24 | 15 | 501 | 1 | 1 | ||
| B | Low | Low | – | 32 | 24 | 407 | 1.272 (0.664–2.435) | 0.469 | 1.229 (0.541–2.796) | 0.622 |
| C | High | Low | – | 32 | 15 | 883 | 3.533 (1.730–7.215) | 0.001 | 4.598 (1.948–10.851) | <0.001 |
| D | High | High | – | 24 | 15 | 592 | 1.549 (0.786–3.055) | 0.206 | 1.797 (0.782–3.682) | 0.181 |
| I | Low | – | High | 25 | 17 | 521 | 1 | 1 | ||
| II | Low | – | Low | 31 | 22 | 373 | 0.531 (0.275–1.025) | 0.057 | 0.511 (0.232–1.129) | 0.097 |
| III | High | – | Low | 31 | 15 | 826 | 2.221 (1.063–4.639) | 0.034 | 3.294 (1.265–8.574) | 0.015 |
| IV | High | – | High | 25 | 15 | 652 | 1.269 (0.611–2.635) | 0.523 | 1.216 (0.530–2.791) | 0.645 |
| 1 | – | Low | Low | 42 | 21 | 773 | 1 | 1 | ||
| 2 | – | Low | High | 14 | 9 | 630 | 0.671 (0.303–1.484) | 0.324 | 0.705 (0.273–1.819) | 0.47 |
| 3 | – | High | Low | 14 | 11 | 506 | 0.546 (0.257–1.159) | 0.115 | 0.495 (0.215–1.139) | 0.098 |
| 4 | – | High | High | 42 | 28 | 509 | 0.442 (0.245–0.797) | 0.007 | 0.448 (0.225–0.891) | 0.022 |
Italic P values indicate statistically significant. a, adjusted for histologic grade, radiation therapy, radical resection, and targeted molecular therapy. SLC, solute carrier; MST, median survival time; OS, overall survival; PDAC, pancreatic ductal adenocarcinoma; HR, hazard ratio; CI, confidence interval.
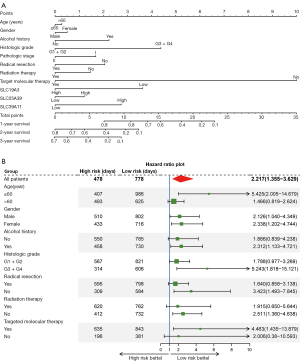
The prognostic signature model, which included SLC19A3, SLC25A39, and SLC39A11 for OS is shown in Figure 5A. The prognosis of the high-risk group was significantly worse than that of the low-risk group (778 vs. 478 days; P=0.001). The risk score, survival status, and heatmap are presented from top to bottom. The ROC curves for predicting the OS of PDAC patients according to SLCs and risk score showed diagnostic value. The area under the curve (AUC) of SLC19A3 for predicting the 1-, 2-, and 3-year survival was 62.2%, 73.3%, and 74.1%, respectively, which was the highest among the genes (Figure 5B).
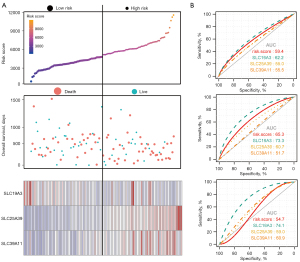
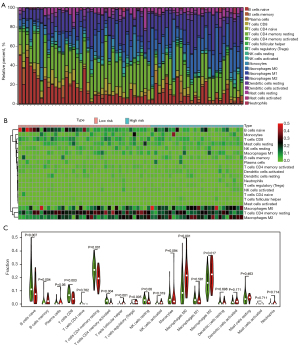
The situation of immune infiltration
After CIBERSORT calculations, 63 out of 112 PDAC samples with P values <0.05 were included in the subsequent analyses. Since the percentage of eosinophils and gamma delta T cells could not be counted, 20 types of immune cells in 63 PDAC samples were analyzed. The proportion of 20 types of immune cells in each sample and a heat map of the immune cells are presented in Figure 6A,6B. Naïve B cells and M2 macrophages were moderately negatively correlated. The proportion of memory B cells (P=0.004), naive B cells (P=0.007), CD8 T cells (P=0.003), activated memory CD4 T cells (P=0.004), and activated NK cells (P=0.019) were significantly higher in the low-risk group compared to that of the high-risk group. In contrast, the proportion of follicular helper T cells (P<0.001), regulatory T cells (Tregs) (P=0.005), M0 macrophages (P<0.001), and M2 macrophages (P=0.017) were higher in the high-risk group (Figure 6C). The Pearson’s correlation coefficient was presented in the matrix box of the correlation graph, and activated natural killer (NK) cells and regulatory T cells (Tregs), naïve CD4 T cells, and memory B cells constituted the median and were moderately positively correlated (Figure 7).
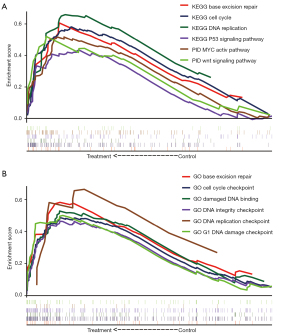
GSEA
In the ROC curves, SLC19A3 was the most eye-catching gene with a high AUC. Therefore, the potential mechanisms associated with the level of SLC19A3 expression on the prognosis of patients with pancreatic cancer were explored in GSEA. In the c2 gene set, the low SLC19A3 expression group was enriched for genes in the cell cycle, base excision repair, DNA replication, MYC active pathway, WNT signaling pathway, and p53 signaling pathway (Figure 8A). The c5 gene set results suggested that base excision repair, cell cycle checkpoint, damaged DNA binding, DNA replication, DNA damage checkpoint, and DNA integrity checkpoint were the main associated functions (Figure 8B).
Discussion
In this study, we performed both a survival and bioinformatic analysis using high-throughput RNA-sequencing data obtained from TCGA. In the survival analysis, the expression of SLC19A3, SLC25A39, and SLC39A11 was found to have the ability to significantly affect the prognosis of PDAC patients; however, the expression of SLC19A3 was completely opposite to that of SLC25A39 and SLC39A11, and overexpression led to a better patient prognosis. This finding is also consistent with previous research. In particular, SLC5A8 also acted as a tumor suppressor gene, and SLC7A11 inhibited pancreatic carcinoma via the PI3K/Akt signaling pathway (39,40). However high SLC28A1 expression indicated a worse prognosis, and SLC22A3 and SLC29A3 affected the therapeutic effect of nucleoside drugs in PC (41). Due to the substantial heterogeneity of both tumors and individuals, a single gene often cannot reflect the effect of the level of gene expression on the prognosis of PDAC. Hence, we constructed a prognostic signature and genetic risk score model to assess the prognosis. In addition, the forest plot showed some differences in clinical factors and patient sensitivity to certain treatment modalities. For example, in the context that radiotherapy can indeed prolong the OS of patients, patients in the low-risk group did not receive radiotherapy, and the OS was similar to those who received radiotherapy. Therefore, we believe that our prognostic signature could predict the prognosis and also provide some evidence for clinical decision-making. In our ROC curves and nomogram, all 3 genes showed excellent efficacy for predicting the prognosis, of which SLC19A3 was the most remarkable. Therefore, we inferred that the 3 genes were independent prognostic factors for PDAC patients.
In most of the previous research, variation of SLC19A3 lead to a thiamine deficiency and resulted in certain genetic disorders, including biotin thiamine responsive basal ganglia disease (BTRBGD), Leigh syndrome, and mitochondrial disorders (42,43). Thiamine transporter-2 (ThTR-2, encoded by SLC19A3) is a specific transporter that plays a role in small intestinal absorption and cellular uptake when thiamine is a critical cofactor for nucleic acid synthesis (44,45). These conclusions are also consistent with our GSEA findings that SLC19A3 participates in the cell cycle, DNA replication, and DNA damage checkpoints. In addition, diabetes is a proven risk factor for PC, and metformin is an effective oral medicine. A study conducted by Liang et al. reported that metformin was a substrate of thiamine transporters and that inhibiting ThTR-2 could reduce the uptake of metformin (5,46). Similarly, cigarette smoking is also a well-known risk factor. Srinivasan et al. reported that chronic nicotine exposure inhibited the uptake of thiamin, as well as the level of ThTR-2 and SLC19A3 expression in pancreatic acinar cells (47). Furthermore, a breast cancer (BC) study performed by Liu et al. reported that the down-regulation of gene expression contributes to the resistance of tumor cells to apoptosis (48). The above results indicate that the level of SLC19A3 expression can affect prognosis and is also associated with the risk factors of PC. Thus, SLC19A3 could act as a new tumor suppressor in PDAC.
We also investigated the potential mechanism of SLC19A3 on PC by GSEA. The results showed that the expression of SLC19A3 was associated with cell cycle, DNA replication, DNA damage, and integrity checkpoints. These findings indicate that SLC19A3 may be involved in the transportation of substances during DNA synthesis and replication to ensure normal gene replication. In addition, the low expression of SLC19A3 is enriched in the MYC and p53 signaling pathways, which are considered the key mutation signaling pathways in PC (49,50).
Mutation of p53 is one of the most common genetic mutations in tumors, and it has been reported that p53 is missing or mutated in approximately 75% of PDAC patients (51). In a mouse model of PDAC, the presence of KRAS and the loss of p53 resulted in a loss in autophagy that did not stop tumor progression but actually accelerated tumor progression (52). Therefore, a lack of SLC19A3 was associated with the p53 signaling pathway, and finally resulted in the loss of autophagy and progression of PDAC.
Some researchers have demonstrated that transcriptomic analysis can be used to explore the immune infiltration microenvironment (53). Tumor-recruited M2 macrophages promote gastric and breast cancer metastasis via M2 macrophage secreted CHI3L1 protein, and it was one the mechanism of M2 macrophages leading to poor prognosis (54). CD8 T cells, CD4 T cells, and NK cells could inhibit tumor progression and metastasis through cytotoxic effects (55). Different types of immune cells also have different effects on tumor progression. For example, cytotoxic CD8 T cells and CD4 helper T cells can inhibit tumor progression, and a high level of activated CD8 T cells can prolong the prognosis of patients (56). In contrast, macrophages, mast cells, and neutrophils can promote tumor progression and are not conducive to the patient’s prognosis (56). We analyzed the situation of TIICS in both high- and low-risk groups using CIBERSORT and found that the TICCs favorable to prognosis (e.g., CD8 T cells, CD4 T cells, and NK cells) were significantly higher in the low-risk group. Simultaneously, M0 and M2 macrophages, which have been found to be less favorable for prognosis were significantly higher in the high-risk group. Immunotherapy including immune checkpoint blockade is not effective in PDAC (57). Therefore, there is an urgent need for new immunotherapy targets to improve the prognosis of patients in PDAC. A recent report demonstrated that the selective targeting of MHC-I molecules for degradation could enhance autophagy and leads to an improved therapeutic strategy (58). It is known that autophagy and apoptosis are indispensable steps in immunotherapy, as well as the suppression of tumorigenesis and progression. Moreover, some SLCs have been shown to participate in glucose uptake and lactate release, as well as the promotion phagocyte engulfment of apoptotic cells (59). In addition, SLC genes can modify dendritic cells and induce an anti-gastric cancer immune response (60). Therefore, SLCs may be a potential target of PDAC immunotherapy, which can promote patient prognosis in the high-risk patient group.
Previous studies on SLCs have suggested that the level of SLC expression is related to the prognosis of certain tumors. For example, SLC39A7, SLC39A11, and SLC39A14 have been associated with the prognosis in GC, and the co-expression of SLC1A5, SLC7A5, and SLC3A2 has been shown to affect aggressive BC which is driven by c-MYC (61,62). However, we first pointed out that SLC19A3, SLC25A39, and SLC39A11 were significantly associated with the prognosis of PDAC, and the potential mechanisms were explored. A prognostic signature was proposed when immunotherapy did not have a good effect on PDAC, and it was calculated that TIICS differed between the high- and low-risk groups. These findings are promising for providing new targets for treatment and biomarkers for predicting the prognosis of PDAC patients.
This study had several limitations. First, the analysis involved publicly available data, and thus, information for some patients was missing (e.g., the patient’s medication, and immune-related medical history), which prevented us from performing further research on these patients. Second, although we established strict inclusion and exclusion criteria, our research was based on a bioinformatics analysis. Therefore, the obtained results still require further experimental verification. Third, due to the difficulty in obtaining PDAC tumor tissue, the level of messenger (mRNA) expression for some SLCs was derived from a single cohort, and a verification cohort was lacking. Fourth, in order to enhance the credibility of these results, we established strict standards, which also led to a small number of samples. A larger sample size is required to minimize bias in future studies.
Despite these limitations, this was the first study to report that SLC19A3, SLC25A35, and SLC39A11 and the prognosis of PDAC patients are significantly correlated. The mechanism by which SLC19A3 expression affects PDAC and the differences in tumor infiltrating cells within the tumor were explored using GSEA and CIBERSORT, respectively. Upon future confirmation of these conclusions, SLC19A3 may represent a new PDAC tumor suppressor that can contribute to the management and treatment of PDAC patients.
Conclusions
The SLC19A3, SLC25A35, and SLC39A11 genes are significantly associated with the prognosis of PDAC patients and may act as a potential biomarker for predicting the prognosis of PDAC following a pancreaticoduodenectomy. The SLC19A3 gene may represent a tumor suppressor in PDAC and affect tumor development and progression through the MYC and p53 signaling pathways and changes in immune cell infiltration. However, these findings should be verified through future functional experiments.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6341/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6341/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lin QJ, Yang F, Jin C, et al. Current status and progress of pancreatic cancer in China. World J Gastroenterol 2015;21:7988-8003. [Crossref] [PubMed]
- Raimondi S, Maisonneuve P, Lowenfels AB. Epidemiology of pancreatic cancer: an overview. Nat Rev Gastroenterol Hepatol 2009;6:699-708. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Gillen S, Schuster T, Meyer Zum Büschenfelde C, et al. Preoperative/neoadjuvant therapy in pancreatic cancer: a systematic review and meta-analysis of response and resection percentages. PLoS Med 2010;7:e1000267. [Crossref] [PubMed]
- Kamisawa T, Wood LD, Itoi T, et al. Pancreatic cancer. Lancet 2016;388:73-85. [Crossref] [PubMed]
- Tang K, Lu W, Qin W, et al. Neoadjuvant therapy for patients with borderline resectable pancreatic cancer: A systematic review and meta-analysis of response and resection percentages. Pancreatology 2016;16:28-37. [Crossref] [PubMed]
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9-29. [Crossref] [PubMed]
- Schaller L, Lauschke VM. The genetic landscape of the human solute carrier (SLC) transporter superfamily. Hum Genet 2019;138:1359-77. [Crossref] [PubMed]
- Cedernaes J, Olszewski PK, Almén MS, et al. Comprehensive analysis of localization of 78 solute carrier genes throughout the subsections of the rat gastrointestinal tract. Biochem Biophys Res Commun 2011;411:702-7. [Crossref] [PubMed]
- Mäkelä S, Kere J, Holmberg C, et al. SLC26A3 mutations in congenital chloride diarrhea. Hum Mutat 2002;20:425-38. [Crossref] [PubMed]
- Al-Suyufi Y. SLC5A1 Mutations in Saudi Arabian Patients With Congenital Glucose-Galactose Malabsorption. J Pediatr Gastroenterol Nutr 2018;66:250-2. [Crossref] [PubMed]
- Cannizzaro M, Jarošová J, De Paepe B. Relevance of solute carrier family 5 transporter defects to inherited and acquired human disease. J Appl Genet 2019;60:305-17. [Crossref] [PubMed]
- Li H, Myeroff L, Smiraglia D, et al. SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proc Natl Acad Sci U S A 2003;100:8412-7. [Crossref] [PubMed]
- Gamble LD, Purgato S, Murray J, et al. Inhibition of polyamine synthesis and uptake reduces tumor progression and prolongs survival in mouse models of neuroblastoma. Sci Transl Med 2019;11:eaau1099. [Crossref] [PubMed]
- Girardi E, César-Razquin A, Lindinger S, et al. A widespread role for SLC transmembrane transporters in resistance to cytotoxic drugs. Nat Chem Biol 2020;16:469-78. [Crossref] [PubMed]
- Lemstrová R, Souček P, Melichar B, et al. Role of solute carrier transporters in pancreatic cancer: a review. Pharmacogenomics 2014;15:1133-45. [Crossref] [PubMed]
- Sprowl JA, Ness RA, Sparreboom A. Polymorphic transporters and platinum pharmacodynamics. Drug Metab Pharmacokinet 2013;28:19-27. [Crossref] [PubMed]
- Zhou J, Hui X, Mao Y, et al. Identification of novel genes associated with a poor prognosis in pancreatic ductal adenocarcinoma via a bioinformatics analysis. Biosci Rep 2019;39:BSR20190625. [Crossref] [PubMed]
- Schulz M, Salamero-Boix A, Niesel K, et al. Microenvironmental Regulation of Tumor Progression and Therapeutic Response in Brain Metastasis. Front Immunol 2019;10:1713. [Crossref] [PubMed]
- Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19:1423-37. [Crossref] [PubMed]
- Fan JQ, Wang MF, Chen HL, et al. Current advances and outlooks in immunotherapy for pancreatic ductal adenocarcinoma. Mol Cancer 2020;19:32. [Crossref] [PubMed]
- Robert C, Long GV, Brady B, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med 2015;372:320-30. [Crossref] [PubMed]
- Ansell SM, Lesokhin AM, Borrello I, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med 2015;372:311-9. [Crossref] [PubMed]
- Kang YK, Boku N, Satoh T, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;390:2461-71. [Crossref] [PubMed]
- Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455-65. [Crossref] [PubMed]
- Group Young Researchers In Inflammatory Carcinogenesis. Wandmacher AM, Mehdorn AS, Sebens S. The Heterogeneity of the Tumor Microenvironment as Essential Determinant of Development, Progression and Therapy Response of Pancreatic Cancer. Cancers (Basel) 2021;13:4932. [Crossref] [PubMed]
- Kiryu S, Ito Z, Suka M, et al. Prognostic value of immune factors in the tumor microenvironment of patients with pancreatic ductal adenocarcinoma. BMC Cancer 2021;21:1197. [Crossref] [PubMed]
- Ansari RE, Craze ML, Althobiti M, et al. Enhanced glutamine uptake influences composition of immune cell infiltrates in breast cancer. Br J Cancer 2020;122:94-101. [Crossref] [PubMed]
- Yip SH, Wang P, Kocher JA, et al. Linnorm: improved statistical analysis for single cell RNA-seq expression data. Nucleic Acids Res 2017;45:e179. [Crossref] [PubMed]
- Liao X, Huang K, Huang R, et al. Genome-scale analysis to identify prognostic markers in patients with early-stage pancreatic ductal adenocarcinoma after pancreaticoduodenectomy. Onco Targets Ther 2017;10:4493-506. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214-20. [Crossref] [PubMed]
- Gentles AJ, Newman AM, Liu CL, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 2015;21:938-45. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Liberzon A, Birger C, Thorvaldsdóttir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417-25. [Crossref] [PubMed]
- François O, Martins H, Caye K, et al. Controlling false discoveries in genome scans for selection. Mol Ecol 2016;25:454-69. [Crossref] [PubMed]
- Park JY, Helm JF, Zheng W, et al. Silencing of the candidate tumor suppressor gene solute carrier family 5 member 8 (SLC5A8) in human pancreatic cancer. Pancreas 2008;36:e32-9. [Crossref] [PubMed]
- Zhu JH, De Mello RA, Yan QL, et al. MiR-139-5p/SLC7A11 inhibits the proliferation, invasion and metastasis of pancreatic carcinoma via PI3K/Akt signaling pathway. Biochim Biophys Acta Mol Basis Dis 2020;1866:165747. [Crossref] [PubMed]
- Mohelnikova-Duchonova B, Brynychova V, Hlavac V, et al. The association between the expression of solute carrier transporters and the prognosis of pancreatic cancer. Cancer Chemother Pharmacol 2013;72:669-82. [Crossref] [PubMed]
- Pronicka E, Piekutowska-Abramczuk D, Ciara E, et al. New perspective in diagnostics of mitochondrial disorders: two years’ experience with whole-exome sequencing at a national paediatric centre. J Transl Med 2016;14:174. [Crossref] [PubMed]
- Ortigoza-Escobar JD, Alfadhel M, Molero-Luis M, et al. Thiamine deficiency in childhood with attention to genetic causes: Survival and outcome predictors. Ann Neurol 2017;82:317-30. [Crossref] [PubMed]
- Asencio C, Rodríguez-Hernandez MA, Briones P, et al. Severe encephalopathy associated to pyruvate dehydrogenase mutations and unbalanced coenzyme Q10 content. Eur J Hum Genet 2016;24:367-72. [Crossref] [PubMed]
- Ah Mew N, Loewenstein JB, Kadom N, et al. MRI features of 4 female patients with pyruvate dehydrogenase E1 alpha deficiency. Pediatr Neurol 2011;45:57-9. [Crossref] [PubMed]
- Liang X, Chien HC, Yee SW, et al. Metformin Is a Substrate and Inhibitor of the Human Thiamine Transporter, THTR-2 (SLC19A3). Mol Pharm 2015;12:4301-10. [Crossref] [PubMed]
- Srinivasan P, Thrower EC, Loganathan G, et al. Chronic Nicotine Exposure In Vivo and In Vitro Inhibits Vitamin B1 (Thiamin) Uptake by Pancreatic Acinar Cells. PLoS One 2015;10:e0143575. [Crossref] [PubMed]
- Liu S, Huang H, Lu X, et al. Down-regulation of thiamine transporter THTR2 gene expression in breast cancer and its association with resistance to apoptosis. Mol Cancer Res 2003;1:665-73. [PubMed]
- English IA, Sears RC. Deconstructing Pancreatic Adenocarcinoma by Targeting the Conductor, MYC. Cancer Discov 2020;10:495-7. [Crossref] [PubMed]
- Ogawa S, Fukuda A, Matsumoto Y, et al. SETDB1 Inhibits p53-Mediated Apoptosis and Is Required for Formation of Pancreatic Ductal Adenocarcinomas in Mice. Gastroenterology 2020;159:682-696.e13. [Crossref] [PubMed]
- Nigro JM, Baker SJ, Preisinger AC, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature 1989;342:705-8. [Crossref] [PubMed]
- Rosenfeldt MT, O’Prey J, Morton JP, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature 2013;504:296-300. [Crossref] [PubMed]
- Teschendorff AE, Miremadi A, Pinder SE, et al. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol 2007;8:R157. [Crossref] [PubMed]
- Chen Y, Zhang S, Wang Q, et al. Tumor-recruited M2 macrophages promote gastric and breast cancer metastasis via M2 macrophage-secreted CHI3L1 protein. J Hematol Oncol 2017;10:36. [Crossref] [PubMed]
- Ravindranath MH, Filippone EJ, Devarajan A, et al. Enhancing Natural Killer and CD8+ T Cell-Mediated Anticancer Cytotoxicity and Proliferation of CD8+ T Cells with HLA-E Monospecific Monoclonal Antibodies. Monoclon Antib Immunodiagn Immunother 2019;38:38-59. [Crossref] [PubMed]
- Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298-306. [Crossref] [PubMed]
- O’Reilly EM, Oh DY, Dhani N, et al. Durvalumab With or Without Tremelimumab for Patients With Metastatic Pancreatic Ductal Adenocarcinoma: A Phase 2 Randomized Clinical Trial. JAMA Oncol 2019;5:1431-8. [Crossref] [PubMed]
- Yamamoto K, Venida A, Yano J, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 2020;581:100-5. [Crossref] [PubMed]
- Morioka S, Perry JSA, Raymond MH, et al. Efferocytosis induces a novel SLC program to promote glucose uptake and lactate release. Nature 2018;563:714-8. [Crossref] [PubMed]
- Xue G, Cheng Y, Ran F, et al. SLC gene-modified dendritic cells mediate T cell-dependent anti-gastric cancer immune responses in vitro. Oncol Rep 2013;29:595-604. [Crossref] [PubMed]
- Ding B, Lou W, Xu L, et al. Analysis the prognostic values of solute carrier (SLC) family 39 genes in gastric cancer. Am J Transl Res 2019;11:486-98. [PubMed]
- El-Ansari R, Craze ML, Alfarsi L, et al. The combined expression of solute carriers is associated with a poor prognosis in highly proliferative ER+ breast cancer. Breast Cancer Res Treat 2019;175:27-38. [Crossref] [PubMed]
(English Language Editor: J. Jones)



