
The regulatory roles of miR-26a in the development of fracture and osteoblasts
Introduction
With the aging of the population becoming more and more serious, osteoporosis (OP) has become a major public health problem worldwide (1,2). OP is a systemic bone disease which leads to decreased bone mineral density and bone mass, and damaged microstructures (3). Fractures are the most serious complication of OP, and osteoporotic patients have markedly higher morbidity and mortality due to fractures (4,5).
Under normal physiological conditions, bone is a dynamic tissue consistently and simultaneously reconstructed to maintain volume, microstructure, and strength (6). Bone tissue is composed of several cell types, including osteoblasts, osteoclasts, osteocytes, and bone lining cells (7). These cells are responsible for maintaining the balance of the bone microenvironment (8). When osteoclast-mediated bone resorption predominates over osteoblast-mediated bone formation, bone mass is seriously threatened by gradual loss (9). The pathogenesis of OP is complex. Several reports have shown that the inhibition of bone formation and the activation of bone resorption in a microgravity environment are the main causes for OP (10). Furthermore, the suppression of bone formation results from a decrease in osteoblast activity (11). It has also been reported that the apoptosis of osteoblasts leads to the pathogenesis of hormone-induced femoral head necrosis, which is regarded as the cytological basis of necrosis of the femoral head (12). Osteoblast dysfunction is considered as the main cause of OP-related bone loss. However, the cellular mechanisms underlying changes in osteoblast function remain unclear.
Among the factors that can alter gene expression are microRNAs (miRNAs) (13,14). MiRNAs are a class of small, single-stranded, non-coding RNAs of about 22 nucleotides or less in length (8,15,16). MiRNAs have been reported to be highly conserved in many species and participate in the regulation of many biological processes (17), mainly by negatively regulating the translation of their target mRNAs (18). MiRNAs exert post-transcriptional control via inhibiting or degrading the target genes. Their functions refer to both physiological and pathological conditions, including metabolism, differentiation, apoptosis, and various diseases (19). Their roles in skeletal development have been investigated by several studies (20). Recently, many studies have demonstrated that miRNAs play vital roles in bone development. For example, some miRNAs are able to positively or negatively regulate the function of osteoblasts, including miR-3077-5p, miR-3090-5p, miR-3103-5p, miR-466i-3p, and miR-466h-3p (21,22).
This study will investigate the role of miRNAs in skeletal development, mainly in fractures, and clarify the miRNAs that are critical factors for skeletal development. We found that the direct effects of miR-26a are to regulate the cell viability, proliferation, and apoptosis of osteoblasts. The indirect effect of miR-26a is to restore bone mass by elevating the functions of osteoblasts, affecting bone formation in the bone microenvironment. Thus, our study has theoretical and clinical significance in preventing the occurrence of bone diseases, including OP and fracture.
We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6101/rc).
Methods
Clinical sample collection
From May 2018 to January 2020, 8 samples of blood samples were collected from patients with fractures and another 8 blood samples were obtained from healthy volunteers who were admitted to the First Affiliated Hospital of Harbin Medical University. These samples were used to analyze the expression levels of miRNAs between healthy controls and patients with fractures.
All participants were provided with detailed information, including the clinical, pathological, and prognostic aspects, and were diagnosed by at least two clinical physicians. The present study was approved by the Ethics Committee of the First Affiliated Hospital of Harbin Medical University (No. 2020037). All the subjects signed a written informed consent form and participated in our study willingly. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture
Human osteoblasts hFOB1.19 were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Osteoblasts were cultured in α-MEM medium (Hyclone, USA) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc., USA), 100 U/mL penicillin, and 100 μg/mL streptomycin, and placed in a 5% CO2 humidified incubator (Thermo Fisher Scientific, Inc., USA) at 37 °C. When the confluence reached approximately 90%, the cells were subcultured to the next passage. The medium was changed every 3 days.
Cell transfection
Transfection with miRNAs was performed using X-treme (Vazyme, China) according to the manufacturer’s instructions. Details of the experimental designs are shown below. All the mimics, inhibitors, and negative controls (NCs) for miR-26a were designed and synthesized by Genepahrma (Shanghai, China). Cells were divided into the following four groups: miR-26a mimics, mimics NC, miR-26a inhibitor, and inhibitor NC group. The final concentration of miR-26a mimics or mimics NC transfected into cells was 50 nM. The final concentration of miR-26a inhibitor or inhibitor NC transfected into cells was 100 nM. Then, the cells were cultured in fresh medium supplemented with 10% FBS to maintain the normal growth of osteoblasts.
MTT assay
The cell viability of osteoblasts was measured by the MTT assay. Briefly, approximately 1×104 human osteoblasts were seeded into each well of a 96‑well plate and placed in an incubator. When the cells grew up to 50–60% density, miRNAs were transfected into the cells. After 0, 12, 24, 36, and 48 h, the MTT assay was performed. The absorbance of human osteoblasts at 490 nm was detected by a microplate reader (TECAN, Switzerland).
EdU staining
EdU staining assay was applied by using EdU Apollo567 Kit (Ribobio, China) following the instructions. The pictures of images of the staining were taken by using a fluorescent microscope (Olympus, Japan). The red color indicated the proliferative cells and the green color indicated the number of total cells.
TUNEL staining
The osteoblasts were transfected with miR-26a mimics, miR-26a inhibitor, and their corresponding controls for 24 h. After transfection, the cells were harvested and washed with PBS at least 3 times. After washing, the cells were fixed in 4% PFA for 20 min at room temperature and TUNEL staining was performed. The cells were incubated with a terminal deoxynucleotidyl transferase‑mediated dUTP nick-end labeling assay kit (Roche, Switzerland). Finally, the apoptotic cells were detected and analyzed under a microscope (Nikon, Japan).
Trypan blue dye exclusion assay
Cell death was assessed by performing the trypan blue dye exclusion assay based on the manufacturer’s instructions. In brief, cells were seeded in 6-well plates and transfected with miR-26a. After 24 h, the osteoblasts were trypsinized using 500 μL of trypsin (Beyotime, China) and the mixture of detached osteoblasts was rinsed in PBS. After washing, the mixture was centrifuged at 1,200 rpm for 3 min. Next, the residue was treated with 500 μL 0.4% trypan blue solution. After the cells were stained, the cells were analyzed using an automated cell counter (TC10, BioRad). Dead cells exhibited a blue color and live cells were not able to be stained with the blue dye. Cell death (%) was counted and analyzed.
Alizarin red S (ARS) staining
ARS staining was used to detect the mineralization deposits of osteoblasts. Osteoblasts were plated at a density of approximately 2×104 cells per well in 24-well plates and cultured in osteogenic differentiation induced medium composed of 10% FBS, 1% glutamine, 1% penicillin-streptomycin,0.2% ascorbic acid, 1% β-glycerophosphate, and 0.01% dexamethasone for 10 days. After 10 days, the cells were fixed in 4% PFA and washed with PBS twice. Then, the cells were stained with ARS staining solution (Cyagen, USA) for 20 min. The stained matrix was observed under a microscope (Nikon, Japan).
Quantitative real-time polymerase chain reaction (qRT-PCR) assay
Total RNAs were extracted from blood samples and cell samples using the TRIzol reagent (Thermo Fisher Scientific, Inc., USA) on the basis of the manufacturer’s protocol (23). In brief, RNAs were reverse-transcribed to cDNAs using a reverse transcription kit (Applied bioscience, USA). Total RNAs (0.5 μg), double distilled water (ddH2O), and reverse transcription reagents were mixed and used for the reverse transcription reaction. Subsequently, qRT-PCR was performed using a SYBR kit (Takara Bio, Japan), and U6 was used as the reference gene to normalize the expression levels of target genes. The PCR reaction (20 μL) system contained 10 μL SYBR reagent, 1 μL cDNA, 2 μL primers, and 7 μL ddH2O. The thermocycler conditions were: 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 15 sec and an annealing/elongation step at 60 °C for 30 sec. The relative expression of target genes was analyzed using the 2–ΔΔCt method. The following primers (Genepharma, China) were used for qRT-PCR detection: U6 (forward: 5'-AGAGAAGATTAGCATGGCCCCTG-3', reverse: 5'-ATCCAGTGCAGGGTCCGAGG-3'); miR-218 (forward: 5'-TTGCGGATGGTTCCGTCAAGCA-3', reverse: 5'-ATCCAGTGCAGGGTCCGAGG-3'); miR-26a (forward: 5'-TTGGATCCGTCAGAAATTCTCTCCCGAGG-3', reverse: 5'-GGTCTAGATGTGAACTCTGGTGTTGGTGC-3'); miR‑181a (forward: 5'‑AACATTCAACGCTGTCGGTGAGT‑3', reverse: 5'‑CTCCTTAGAATCTGTTTGCTCTCATA‑3'); miR-132-3p (forward: 5'-GCGCGCGTAACAGTCTACAGC-3', reverse: 5'-GTCGTATCCAGTGCAGGGTCC-3'); miR-638 (forward: 5'-AGGGATCGCGGGCGGGT-3', reverse: 5'-CAGTGCAGGGTCCGAGGT-3'); alkaline phosphatase (ALP) (forward: 5'-ACAACCTGACTGACCCTTCG-3', reverse: 5'-TCATGATGTCCGTGGTCAAT-3'); bone morphogenetic protein 4 (BMP4) (forward: 5'-TCGTTACCTCAAGGGAGTGG-3', reverse: 5'-ATGCTTGGGACTACGTTTGG-3'); osteocalcin (OCN) (forward: 5'-TTCTGCTCACTCTGCTGACC-3', reverse: 5'-TTTGTAGGCGGTCTTCAAGC-3').
Dual luciferase reporter assay
Dual luciferase activity assay was performed to identify whether phosphatase and tensin homolog (PTEN) was a target of miR-26a. The binding sites for miR-26a in the 3'-UTR of PTEN was constructed and cloned into the pmirGLO dual luciferase vector, which was named as pmirGLO-PTEN-WT. The mutated 3'-UTR sequences of PTEN were also cloned, which was named as pmirGLO-PTEN-MUT. Then, HEK-293T cells were seeded in 24-well plates (Corning, USA) and transfected with 100 ng of empty vector, pmirGLO-PTEN-WT, or pmirGLO-PTEN-MUT together with miR-26a using Lipofectamine 2000 (Invitrogen, USA). After 24 h, the dual luciferase activity was detected by using dual luciferase reporter assay kit (Promega, USA).
Western blot assay
The western blotting analysis was performed according to standard methods. In brief, total proteins of cell lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes. Then, the membranes were blocked by 5% non-fat milk and treated with primary antibodies (and β-actin) overnight at 4 °C. Then, membranes were incubated with secondary antibody at room temperature for 2 h. After washed by TBST, the protein blots were visualized using an enhanced chemiluminescence kit (Santa, China) and Image J software (NIH, USA) was used to quantify the intensity of each band.
Statistical analysis
All the experiments were conducted at least 3 times. Data were presented as the mean ± standard deviation (SD). Analysis in our study was performed using Graphpad prism software (Graphpad, USA). Comparisons between groups were made using the Student’s t-test or one-way multivariate analysis of variance (ANOVA). P<0.05 was considered to indicate a statistically significant difference.
Results
The expression of miR-26a in patients with fractures
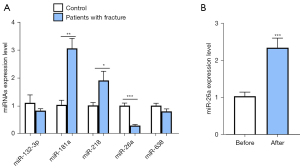
First, through searching multiple miRNA databases and reports, we found that some miRNAs, including miR-132-3p, miR-181a, miR-218, miR-26a, and miR-638, are involved in bone development. However, there are no reports that have investigated whether these miRNAs are related to the development of fractures. To determine the expression patterns of miRNAs in patients with fractures, 8 pairs of blood samples from patients with fractures and corresponding healthy volunteers were collected, followed by the detection of the expression levels of miRNAs using qRT-PCR assays. The results showed that the expression levels of miR-132-3p, miR-26a, and miR-638 were down-regulated in patients with fractures. Furthermore, miR-218 and miR-181a were up-regulated in patients with fractures compared with controls (Figure 1A). Among these miRNAs, miR-26a was the most dysregulated miRNA between fracture patients and controls, and the expression of miR-26a in controls was about 5 times higher than that in patients with fractures (Figure 1A). To further investigate the expression of miR-26a, the blood samples of patients with fractures before and after treatment were collected. The results of qRT-PCR analysis indicated that after treatment, the expression of miR-26a was significantly increased (Figure 1B). The results indicated that the expression of miR-26a in patients with fractures was significantly elevated after treatment (Figure 1B). Therefore, we assumed that miR-26a might be involved in the occurrence and development of fractures.
The effects of miR-26a on the cell viability and proliferation of osteoblasts
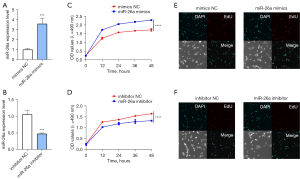
To further determine the potential role of miR-26a in osteoblasts, miR-26a mimics and miR-26a inhibitor were utilized and the transfection efficiency of miR-26a mimics and miR-26a inhibitor was assessed by qRT-PCR analysis. The results of qRT-PCR showed that the expression of miR-26a was also significantly increased after transfection of miR-26a mimics, which suggested that miR-26a was indeed overexpressed in osteoblasts after transfection (Figure 2A). On the other hand, the expression of miR-26a was dramatically down-regulated after transfection of the miR-26a inhibitor, which indicated that miR-26a was successfully knocked down (Figure 2B). Subsequently, we performed MTT assay to assess the cell viability after osteoblasts were transfected with miR-26a mimics. We monitored the cell viability at 0, 12, 24, 36, and 48 h after transfection of miR-26a. As expected, the results of the MTT assay in Figure 2C,2D showed that overexpression of miR-26a markedly increased the cell viability of osteoblasts, while miR-26a inhibitor was able to decrease the cell viability (Figure 2C,2D). EdU staining indicated that overexpression of miR-26a obviously increased the proliferation of osteoblasts, which were decreased by the transfection of miR-26a inhibitor (Figure 2E,2F). These findings indicated that miR-26a elevated the cell viability and proliferation ability of osteoblasts.
The effects of miR-26a on the cell survival rate and apoptosis of osteoblasts
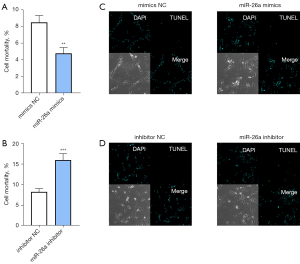
To explore the roles of miR-26a in the cell survival rate and apoptosis of osteoblasts, the cells were transfected with miR-26a mimics, inhibitors, and their corresponding NC, respectively. The trypan blue dye exclusion assay demonstrated that overexpression of miR-26a elevated the cell survival (Figure 3A). In contrast, knockdown of miR-26a decreased the number of live osteoblasts, demonstrating the reduction in cell survival (Figure 3B). Furthermore, miR-26a mimics significantly inhibited the apoptosis of osteoblasts, as evidenced by TUNEL staining (Figure 3C). TUNEL staining showed that knockdown of miR-26a facilitated the apoptosis of osteoblasts, as demonstrated by the increase in the number of apoptotic cells (Figure 3D). Taken together, these data showed that increased miR-26a levels lead to decreases in the cell death and apoptosis of osteoblasts.
The roles of miR-26a in osteoblastic activity
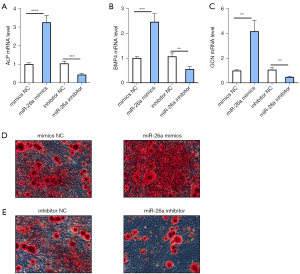
We next examined whether miR-26a regulated the osteoblastic activity. Analysis of osteoblast-associated genes, including ALP, BMP4, and OCN, revealed a significant increase in the levels of these genes after transfection of miR-26a mimics, which promoted osteoblastic activity (Figure 4A-4C). In contrast, downregulation of miR-26a significantly decreased the expression of osteoblast-related genes (Figure 4A-4C). In addition, ARS staining was performed to detect the roles of miR-26a in the mineralized nodules of osteoblasts. Consistent with the qRT-PCR analysis, ARS staining indicated that miR-26a mimics promoted the formation of mineralized nodules in osteoblasts compared to mimics NC (Figure 4D). Furthermore, a reduction of miR-26a in osteoblasts resulted in decreased formation of mineralized nodules (Figure 4E). Taken together, these findings suggested that miR-26a promoted the osteoblastic activity.
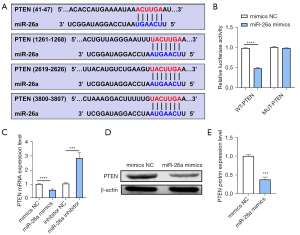
Interaction between miR-26a and PTEN
To analyze the regulatory mechanism of miR-26a, the target genes of miR-26a were predicted online by searching online prediction software TargetScan. The results showed that PTEN, which has been reported to participate in and plays a negative role in the bone regeneration, bone formation and osteogenesis, was a target gene of miR-26a (24-26). And the sequences in the 3'-UTR region of PTEN and miR-26a binding sites were shown in Figure 5A, suggesting miR-26a was bound to the 3'-UTR of PTEN (Figure 5A). Dual luciferase reporter assay was used to analyze the relationship between miR-26a and PTEN, and luciferase activity was assessed in the pmirGLO-PTEN-MUT group and pmirGLO-PTEN-WT group. The results of dual luciferase reporter assay discovered that miR-26a mimic was not able to affect the luciferase activity in the cells co-transfected with pmirGLO-PTEN-MUT, while miR-26a was capable of reducing the luciferase activity in the cells treated with pmirGLO-PTEN-WT group (Figure 5B). Furthermore, qRT-PCR analysis indicated that the mRNA expression of PTEN was reduced by miR-26a overexpression, while increased by knockdown of miR-26a (Figure 5C). In consistent with the results of real time qRT-PCR, western blot analysis also revealed that overexpression of miR-26a suppressed the protein expression level of PTEN (Figure 5D,5E). Therefore, we concluded that miR-26a regulated the biological functions of osteoblasts by targeting its target gene PTEN.
Discussion
The skeleton is comprised of cartilage and bone tissue, and bones undergo constant remodeling throughout our lifetime (27). Bone homeostasis is mainly controlled and regulated by osteoblast-mediated bone formation and osteoclast-mediated bone resorption (28). Therefore, these cell types are vital for the establishment and maintenance of bone remodeling (29). Among them, osteoblasts are a class of bone-forming cells which play a crucial role in bone growth during development and bone formation during remodeling of the postnatal skeleton (13,30). Bone formation depends on the number and activity of osteoblasts in the bone microenvironment (31).
In bones, miRNAs have been reported to regulate a variety of processes, such as the differentiation of osteoblasts and osteoclasts (32,33). Recently, some reports have indicated that miRNAs participate in biological processes such as cancer development, immune regulation, and bone repair (34,35). For example, miR-155 has been shown to inhibit the osteogenic differentiation of mesenchymal stem cells induced by BMP9 via reduction of the BMP signaling pathway (36). According to reports, miR-495 could regulate new bone regeneration and murine femur healing by inhibiting high mobility group AT-Hook 2 (HMGA2) (37). Furthermore, miR-542-3p has been reported to play an important role in bone formation via inhibiting SFRP1 expression and inducing osteoblast differentiation (38). However, the roles of miRNAs in bone remodeling, especially fractures, have not yet been reported. Additionally, the regulatory functions of fracture-related miRNAs in the cell viability, proliferation, apoptosis, cell death, and mineralization of osteoblasts are currently unknown. In addition, through searching multiple miRNA databases and reports, the results showed that some miRNAs, such as miR-132-3p, miR-181a, miR-218, miR-26a, and miR-638, have been reported to participate in the bone development. However, there are no study which analyzed whether these miRNAs are related to the development of fractures.
In our study, among these miRNAs, miR-26a was the most dysregulated miRNA between fracture patients and controls, and the expression of miR-26a in controls was about 5 times higher than that in patients with fractures. Thus, we chose miR-26a for further analysis. In the current study, we unveiled the expression level of miR-26a in patients with fractures. In addition, we detected the roles of miR-26a in the cell viability, proliferation, apoptosis, cell death, and osteoblastic activity of osteoblasts. Based on our evidence, we propose that miR-26a is significantly decreased in patients with fractures, and is associated with the development of fractures through regulating the cell viability, proliferation, apoptosis, cell death, and activity of osteoblasts. The elevation of miR6a in osteoblasts might be a novel strategy for treating fracture involving a reduction in bone formation.
In conclusion, this study is the first to demonstrate the overall importance of miR-26a during skeletal development. Important experiments have revealed a critical role for miR-26a in the cell viability, proliferation, cell death, and apoptosis of osteoblasts. Together, our study has revealed that miR-26a plays an important role in the pathogenesis of fractures, and contributes to the development of new therapeutic approaches for OP.
Acknowledgments
Funding: This work was supported by the Program for Innovation Research of Heilongjiang Provincial Hospital and the Scientific Research Project of Heilongjiang Health Commission (No. 2019-159) and Natural Science Foundation of Heilongjiang Province (No. LH2021H053).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6101/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6101/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6101/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The present study was approved by the Ethics Committee of the First Affiliated Hospital of Harbin Medical University (No. 2020037). All the subjects signed a written informed consent form and participated in our study willingly. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegenthaler B, Ghayor C, Gjoksi-Cosandey B, et al. The Bromodomain Inhibitor N-Methyl pyrrolidone Prevents Osteoporosis and BMP-Triggered Sclerostin Expression in Osteocytes. Int J Mol Sci 2018;19:3332. [Crossref] [PubMed]
- Li Y, Yang F, Gao M, et al. miR-149-3p Regulates the Switch between Adipogenic and Osteogenic Differentiation of BMSCs by Targeting FTO. Mol Ther Nucleic Acids 2019;17:590-600. [Crossref] [PubMed]
- Zhang Z, Chen Y, Xiang L, et al. Diosgenin protects against alveolar bone loss in ovariectomized rats via regulating long non-coding RNAs. Exp Ther Med 2018;16:3939-50. [Crossref] [PubMed]
- Wang Q, Yao L, Xu K, et al. Madecassoside inhibits estrogen deficiency-induced osteoporosis by suppressing RANKL-induced osteoclastogenesis. J Cell Mol Med 2019;23:380-94. [Crossref] [PubMed]
- Yang F, Yang L, Li Y, et al. Melatonin protects bone marrow mesenchymal stem cells against iron overload-induced aberrant differentiation and senescence. J Pineal Res 2017; [Crossref] [PubMed]
- Jiang LB, Tian L, Zhang CG. Bone marrow stem cells-derived exosomes extracted from osteoporosis patients inhibit osteogenesis via microRNA-21/SMAD7. Eur Rev Med Pharmacol Sci 2018;22:6221-9. [PubMed]
- Boyce BF, Li J, Xing L, et al. Bone Remodeling and the Role of TRAF3 in Osteoclastic Bone Resorption. Front Immunol 2018;9:2263. [Crossref] [PubMed]
- Aquino-Martinez R, Farr JN, Weivoda MM, et al. miR-219a-5p regulates Rorβ during osteoblast differentiation and in age-related bone loss. J Bone Miner Res 2019;34:135-44. [Crossref] [PubMed]
- Jiang M, Wang T, Yan X, et al. A novel rhein derivative modulates bone formation and resorption and ameliorates estrogen-dependent bone loss. J Bone Miner Res 2019;34:361-74. [Crossref] [PubMed]
- Li K, Xiu C, Zhou Q, et al. A dual role of cholesterol in osteogenic differentiation of bone marrow stromal cells. J Cell Physiol 2019;234:2058-66. [Crossref] [PubMed]
- Wang H, Hu Z, Shi F, et al. Osteoblast-targeted delivery of miR-33-5p attenuates osteopenia development induced by mechanical unloading in mice. Cell Death Dis 2018;9:170. [Crossref] [PubMed]
- Xi H, Tao W, Jian Z, et al. Levodopa attenuates cellular apoptosis in steroid-associated necrosis of the femoral head. Exp Ther Med 2017;13:69-74. [Crossref] [PubMed]
- Wang Y, Wang K, Hu Z, et al. MicroRNA-139-3p regulates osteoblast differentiation and apoptosis by targeting ELK1 and interacting with long noncoding RNA ODSM. Cell Death Dis 2018;9:1107. [Crossref] [PubMed]
- Li Y, Feng C, Gao M, et al. MicroRNA-92b-5p modulates melatonin-mediated osteogenic differentiation of bone marrow mesenchymal stem cells by targeting ICAM-1. J Cell Mol Med 2019;23:6140-53. [Crossref] [PubMed]
- Yang L, Li Y, Gong R, et al. The Long Non-coding RNA-ORLNC1 Regulates Bone Mass by Directing Mesenchymal Stem Cell Fate. Mol Ther 2019;27:394-410. [Crossref] [PubMed]
- Zou J, Du J, Tu H, et al. Resveratrol benefits the lineage commitment of bone marrow mesenchymal stem cells into osteoblasts via miR-320c by targeting Runx2. J Tissue Eng Regen Med 2021;15:347-60. [Crossref] [PubMed]
- Ramírez-Salazar EG, Carrillo-Patiño S, Hidalgo-Bravo A, et al. Serum miRNAs miR-140-3p and miR-23b-3p as potential biomarkers for osteoporosis and osteoporotic fracture in postmenopausal Mexican-Mestizo women. Gene 2018;679:19-27. [Crossref] [PubMed]
- Pan J, Huang C, Chen G, et al. MicroRNA-451 blockade promotes osteoblastic differentiation and skeletal anabolic effects by promoting YWHAZ-mediated RUNX2 protein stabilization. Medchemcomm 2018;9:1359-68. [Crossref] [PubMed]
- Wang W, Yang L, Zhang D, et al. MicroRNA-218 negatively regulates osteoclastogenic differentiation by repressing the nuclear factor-κB signaling pathway and targeting tumor necrosis factor receptor 1. Cell Physiol Biochem 2018;48:339-47. [Crossref] [PubMed]
- Shao B, Fu X, Yu Y, et al. Regulatory effects of miRNA-181a on FasL expression in bone marrow mesenchymal stem cells and its effect on CD4+T lymphocyte apoptosis in estrogen deficiency-induced osteoporosis. Mol Med Rep 2018;18:920-30. [Crossref] [PubMed]
- Liao L, Yang X, Su X, et al. Redundant miR-3077-5p and miR-705 mediate the shift of mesenchymal stem cell lineage commitment to adipocyte in osteoporosis bone marrow. Cell Death Dis 2013;4:e600. [Crossref] [PubMed]
- Wang Y, Zou X, Guo Y, et al. Mechanical Strain Affects Some Microrna Profiles in Pre-Oeteoblasts. Cell Mol Biol Lett 2015;20:586-96. [Crossref] [PubMed]
- Yang X, Zhang Y, Li Y, et al. MALAT1 enhanced the proliferation of human osteoblasts treated with ultra-high molecular weight polyethylene by targeting VEGF via miR-22-5p. Int J Mol Med 2018;41:1536-46. [Crossref] [PubMed]
- Zheng C, Tang F, Min L, et al. PTEN in osteosarcoma: Recent advances and the therapeutic potential. Biochim Biophys Acta Rev Cancer 2020;1874:188405. [Crossref] [PubMed]
- Chen Y, Yu H, Zhu D, et al. miR-136-3p targets PTEN to regulate vascularization and bone formation and ameliorates alcohol-induced osteopenia. FASEB J 2020;34:5348-62. [Crossref] [PubMed]
- Yang C, Liu X, Zhao K, et al. miRNA-21 promotes osteogenesis via the PTEN/PI3K/Akt/HIF-1α pathway and enhances bone regeneration in critical size defects. Stem Cell Res Ther 2019;10:65. [Crossref] [PubMed]
- Zhou J, Li X, Sun X, et al. Bone regeneration around immediate placed implant of molar teeth with autologous platelet-rich fibrin: Two case reports. Medicine (Baltimore) 2018;97:e13058. [Crossref] [PubMed]
- Wei G, Liang T, Wei C, et al. Daidzin inhibits RANKL-induced osteoclastogenesis in vitro and prevents LPS-induced bone loss in vivo. J Cell Biochem 2019;120:5304-14. [Crossref] [PubMed]
- Wang M, Park S, Nam Y, et al. Bone-fracture-targeted dasatinib-oligoaspartic acid conjugate potently accelerates fracture repair. Bioconjug Chem 2018;29:3800-9. [Crossref] [PubMed]
- Lazić Mosler E, Lukač N, Flegar D, et al. Fas receptor induces apoptosis of synovial bone and cartilage progenitor populations and promotes bone loss in antigen-induced arthritis. FASEB J 2019;33:3330-42. [Crossref] [PubMed]
- Zhang Y, Xing Y, Jia L, et al. An in vitro comparative study of multisource derived human mesenchymal stem cells for bone tissue engineering. Stem Cells Dev 2018;27:1634-45. [Crossref] [PubMed]
- Zhang M, Guo JM, Zhang SH, et al. Research progress on the effect of long non-coding RNA H19 on osteogenic differentiation and bone diseases. Sheng Li Xue Bao 2018;70:531-8. [PubMed]
- Nishino T, Horie T, Baba O, et al. SREBF1/MicroRNA-33b Axis Exhibits Potent Effect on Unstable Atherosclerotic Plaque Formation In Vivo. Arterioscler Thromb Vasc Biol 2018;38:2460-73. [Crossref] [PubMed]
- Qin Y, Tang S, Zhen G, et al. Bone-targeted delivery of TGF-β type 1 receptor inhibitor rescues uncoupled bone remodeling in Camurati-Engelmann disease. Ann N Y Acad Sci 2018;1433:29-40. [Crossref] [PubMed]
- Qiao S, Liu C, Xu W, et al. Up-regulated expression of CD147 gene in malignant bone tumor and the possible induction mechanism during osteoclast formation. Braz J Med Biol Res 2018;51:e6948. [Crossref] [PubMed]
- Liu H, Zhong L, Yuan T, et al. MicroRNA-155 inhibits the osteogenic differentiation of mesenchymal stem cells induced by BMP9 via downregulation of BMP signaling pathway. Int J Mol Med 2018;41:3379-93. [Crossref] [PubMed]
- Tian Z, Zhou H, Xu Y, et al. MicroRNA-495 Inhibits New Bone Regeneration via Targeting High Mobility Group AT-Hook 2 (HMGA2). Med Sci Monit 2017;23:4689-98. [Crossref] [PubMed]
- Zhang X, Zhu Y, Zhang C, et al. miR-542-3p prevents ovariectomy-induced osteoporosis in rats via targeting SFRP1. J Cell Physiol 2018;233:6798-806. [Crossref] [PubMed]


