
The long non-coding RNA rhabdomyosarcoma 2-associated transcript exerts anti-tumor effects on lung adenocarcinoma via ubiquitination of SOX9
Introduction
Lung adenocarcinoma (LUAD) is the most common cancer worldwide. Despite advances in surgical treatment and chemotherapy, the prognosis of LUAD patients is still poor due to metastasis and recurrence. Furthermore, the mechanisms involved in the growth and metastasis of LUAD remain unclear. Understanding the molecular mechanisms underlying the growth and metastasis of LUAD will facilitate the development of improved prognostic indicators and therapeutic targets.
Long noncoding RNAs (lncRNAs) are RNA transcripts with a length of more than 200 nucleotides, that lack important protein coding ability (1). In the past two decades, the mechanisms of microRNAs (miRNAs) and mRNAs have been widely studied. However, the role of lncRNAs remains unclear. LncRNAs can act on tumor stem cells, affecting tumor initiation, development, and metastasis (2). Recently, lncRNAs have been described as ubiquitin-mediated protein hydrolysis inducers and identification of their interacting partners may further our understanding regarding the molecular mechanisms of lncRNAs (3).
Rhabdomyosarcoma 2-associated transcript (RMST) is a lncRNA located in chromosome band 12q23.1.24. Recent studies have reported an association between RMST and glioblastoma multiforme and breast cancer (4,5). Indeed, RMST plays an anti-tumor role in triple negative breast cancer by inhibiting cell proliferation, invasion, and migration. Previous studies have demonstrated that RMST can act as a scaffold to bind different proteins or different protein complexes together (6). For example, RMST interacts with sex determining region Y-box 2 (SOX2) to regulate neurogenesis (6). Peng et al. reported that RMST can enhance DNA methyltransferase 3 (DNMT3) expression through interaction with human antigen R (HuR) (7). However, to date, the expression and function of RMST in LUAD has not been reported.
This study revealed that the expression of RMST was significantly downregulated in LUAD samples compared to healthy controls. Furthermore, RMST suppressed the proliferation and migration of LUAD cells in vitro and in vivo. Notably, RMST was found to directly bind SOX9 to induce its degradation via F-box and WD repeat domain-containing 7 (FBW7). These results suggested that the combination of RMST and SOX9 may be a valuable prognostic factor for LUAD patients.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-6052).
Methods
Tissues and database
Data from The Cancer Genome Atlas (TCGA) and The Genotype-Tissue Expression (GTEx) public databases were used for this study. Furthermore, tissue samples were collected from LUAD patients who underwent surgery in the Zhejiang Provincial People’s Hospital. A total of 90 pairs of tumor and adjacent non-tumor tissues were assessed. Written informed consent was obtained from all patients. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The present study was approved by the Ethics Committee of Zhejiang Provincial People’s Hospital.
Cell culture
The human LUAD cell lines A549 and H1650 were obtained from the American Type Culture Collection (ATCC) and cultured as previously described (8). Cell lines were cultured in RPMI-1640 medium containing 10% fetal bovine serum with 5% CO2 at 37 °C.
Colony formation assay, cell counting kit-8 (CCK8) assays and transwell assay
Colony formation assays, CCK8 assays and transwell assays were performed as previously described (8). In transwell assay, 700 μL RPMI-1640 containing 10% fetal bovine serum was added to the lower chamber, at the same time, 200 μL serum-free medium containing 105 cells was added to the upper chamber. After 24 hours, chambers were fixed in 4% paraformaldehyde and stained with 0.2% crystal violet.
RNA extraction and quantitative polymerase chain reaction (qPCR)
Total RNA was extracted using TRIzol reagent (Invitrogen, USA) and reverse transcribed using the reverse transcription kit from Promega (Madison, USA). The SYBR Green Mix (ABI, CA, USA) was used for qPCR reactions. All primers used in the present study are listed in Table S1.
Western blot analysis
Western blots were performed as previously described (9). The protein was extracted from cell lysate, denatured, transferred to PVDF membrane by electrophoresis, and then incubated with antibody. Antibodies used in this study are listed in Table S2.
Co-immunoprecipitation (Co-IP) assay
Co-IP assays were performed as previously described (10). Cell lysates were incubated with antibodies and magnetic beads before washing and electrophoresis. Antibodies used in this study are listed in Table S2.
Constructs
The sequences of the small interfering RNAs (siRNAs) siRMST, siFBW7, and the negative control (siNC) are listed in Table S3. The RMST overexpression plasmid pcDNA3.1-RMST was purchased from Integrated Biotech Solutions (Shanghai, China).
RNA pull-down assay, mass spectrometry, and RNA immunoprecipitation (RIP) assay
The RiboMAX Large Scale RNA Production Systems (Promega, USA) and the Biotin RNA Labeling Mix (Roche, USA) were used to synthesize full-length RMST in vitro for the RNA pull-down assays. The biotin-labelled RNA was incubated with the cell lysate and the streptavidin-coated beads in accordance with the manufacturer’s instructions. The lysate was then washed three times and examined via Western blot analysis. The protein bands were identified by mass spectrometry.
RIP assays were performed using the RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) according to the manufacturer’s instructions. Briefly, cell lysates were incubated with anti-SOX9 and anti-IgG antibodies. The precipitated RNA was reverse transcribed into cDNA, followed by amplification with RMST primers.
Fluorescence in situ hybridization (FISH)
The FISH Tag RNA Green Kit (Invitrogen, USA) was used according to the manufacturer’s protocol. RMST detection probes were synthesized using linearized plasmids containing T7 RNA polymerase. Hybridization and washing were performed as per the manufacturer’s instructions.
Animal model
Six-week-old male BALB/c nude mice were purchased from the Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China) and housed in specific pathogen-free (SPF) conditions. Mice were randomly assigned to the experimental group and the control group. The LUAD cell lines A549 and H1650 were transfected with plasmids overexpressing RMST or silencing RMST (siRMST), and their respective plasmid negative controls (NC) with Lipofectamine 2000. The transfected cells were subcutaneously injected into mice at a concentration of 2×106 cells/100 µL per mouse. All mice were euthanized at the end of the experiment. The tumor grafts from the mice were harvested and fixed. Tumors were measured and recorded every 5 days, and the tumor volumes were calculated as previously described (11). Animal experiments were performed under a project license [No. KS(Y)21220] granted by ethics board of Zhejiang Provincial People’s Hospital, in compliance with the Guide for the Care and Use of Laboratory Animals of Zhejiang Provincial People’s Hospital. A protocol was prepared before the study without registration.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 5 (La Jolla, CA, USA) and SPSS software version 19.0 (IBM Corporation, Armonk, NY). The experimental data was analyzed using t-tests. A P value <0.05 was considered statistically significant.
Results
Down-regulation of RMST is correlated with progression of lung adenocarcinoma
Sequencing data of LUAD from the TCGA and GTEx databases revealed that RMST expression was downregulated in LUAD samples compared to normal tissues (Figure 1A,1B). In addition, RMST expression was downregulated at all stages of LUAD compared with adjacent healthy tissues (Figure 1C).

The downregulation of RMST in LUAD was validated using clinical samples obtained from patients admitted to our hospital with LUAD (Figure 1D). Kaplan-Meier analysis revealed that low expression of RMST was correlated with poor survival in LUAD patients (Figure 1E).
RMST expression was also detected in different LUAD cell lines. As shown in Figure 1F, A549 cells had the lowest RMST expression, while RMST was highly expressed in H1650 cells. FISH demonstrated that RMST was mostly localized to the nucleus (Figure 1G).
RMST suppresses cell proliferation and migration in vitro and tumorigenicity in vivo
To evaluate the functional role of RMST in LUAD cells, colony formation and cell migration assays were performed. RMST was overexpressed in A549 cells (A549/RMST cells) and knocked down in H1650 cells (H1650/siRMST) and qPCR was used to validate the overexpression and knockdown efficiencies (Figure 2A). Colony formation assays, Cell Counting Kit-8 (CCK8) assays, and transwell assays demonstrated that RMST overexpression in A549 cells resulted in decreased cell proliferation and migration, while RMST knockdown in H1650 accelerated cell proliferation and migration (Figure 2B-2D). These results indicated that RMST suppressed cell proliferation and migration in vitro.
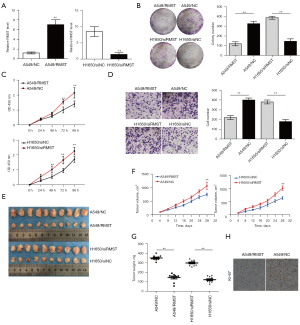
To further validated these effects in vivo, the A549/RMST cells and H1650/siRMST cells were injected into nude mice subcutaneously. Consistent with the in vitro assays, RMST overexpression suppressed tumor volume and weight, while RMST knockdown showed the opposite effect (Figure 2E-2G). The immunohistochemical staining for Ki-67 in tumors harvested form nude mice showed that RMST overexpression resulted in a lower Ki-67 index, while RMST knockdown increased the Ki-67 index (Figure 2H). These results suggested that RMST suppressed tumorigenesis of LUAD cells in vivo.
RMST interacts with SOX9 and promotes SOX9 degradation via ubiquitination
Since lncRNAs can interact with proteins (12), RNA-pull down assays and mass spectrometry (MS) were performed to identify proteins that interact with RMST in LUAD cells. MS analysis revealed that SOX9 was a potential target for interaction with RMST (Figure 3A). RNA-pull down assays and RNA immunoprecipitation (RIP) assays validated the interaction between SOX9 and RMST (Figure 3B,3C). Interestingly, RMST overexpression or knockdown did not alter SOX9 mRNA levels. However, RMST overexpression decreased SOX9 protein levels and RMST knockdown elevated SOX9 protein expression in LUAD cells (Figure 3D,3E). These results suggested that the SOX9 protein might be a target for RMST.

To investigate the mechanisms by which RMST interacts with SOX9, LUAD cells were treated with cycloheximide (CHX). As shown in Figure 3F, RMST overexpression shortened the half-life of the SOX9 protein compared to control cells. Furthermore, when cells were treated with the proteasome inhibitor MG132, SOX9 protein levels were elevated compared with control cells (Figure 3G). These results suggested that ubiquitination may be involved in the RMST-mediated downregulation of SOX9 protein. Indeed, significantly increased levels of ubiquitinated SOX9 protein were detected in cells overexpressing RMST compared to control cells (Figure 3H). Taken together, these results indicated that ubiquitination may be involved in RMST-mediated SOX9 protein degradation.
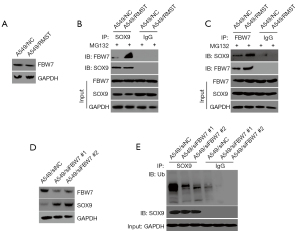
RMST-FBW7 mediates SOX9 ubiquitination
To further investigate the mechanisms by which RMST regulates SOX9 ubiquitination, the mass spectrometry data was analyzed and ubiquitin ligase FBW7 was identified. FBW7 is a ubiquitin ligase component which targets substrates for degradation. As shown in Figure 4A, expression of FBW7 was not altered in cells overexpressing RMST. Indeed, co-IP assays demonstrated that the interaction between SOX9 and FBW7 was strengthened by RMST in A549 cells treated with MG132 (Figure 4B,4C). These data suggested that FBW7 might be a mediator for SOX9 ubiquitination by RMST. When FBW7 expression was silenced in A549 cells, the protein levels of SOX9 increased (Figure 4D). Moreover, SOX9 ubiquitination was significantly decreased in A549/siFBW7 cells compared with negative control cells (Figure 4E). Collectively, these data indicated that RMST may strengthen the interaction between FBW7 and SOX9, and that FBW7 enhances the ubiquitination and degradation of SOX9.
RMST inhibits cell proliferation and migration in a SOX9-dependent manner
Kawai et al. reported that SOX9 can act as a transcriptional factor (TF) (13). To identify the target genes of SOX9, the ChIP-seq data in the public database GSE114305 was analyzed (Figure 5A). The target genes modulated by SOX9 include Smad3, Slug, and ZEB1. These genes are important for inducing the endothelial-mesenchymal transition (EMT) process in various types of tumors (14-16). To determine whether SOX9 modulates Smad3, Slug, and ZEB1 in LUAD, ChIP assays were performed. As shown in Figure 5B, SOX9 directly binds to the promoters of Smad3, Slug, and ZEB1. The PCR results showed that SOX9 knockdown decreased the expression of Smad3, Slug, and ZEB1 (Figure 5C). The protein expression of Smad3, Slug, ZEB1, and other EMT markers were also reduced after silencing of SOX9 (Figure 5D). To evaluate whether RMST functions in LUAD cells in a SOX9-dependent manner, colony-formation and transwell assays were performed. Indeed, RMST silencing increased cell proliferation and migration, and this was impaired by the simultaneous silencing of SOX9 (Figure 5E). Conversely, RMST overexpression inhibited cell proliferation and migration, and this was reversed by the simultaneous overexpression of SOX9 (Figure 5F).

Discussion
Accumulating research has been attracted to lncRNA because of its crucial role in LUAD. Liu et al. reported the relationship between LncRNA and redox (2). Lu et al. reported LncRNA induced ubiquitination of EZH2 (17). An increasing number of studies have focused on the post-translational modification of proteins by lncRNA, such as ubiquitination. Evidence shows that ubiquitination modification plays an important role in tumorigenesis. As an important post-translational modification of proteins, ubiquitination has always been the focus and hotspot of tumor biology research. Ubiquitination modification is a reversible enzyme cascade reaction, which is precisely regulated by ubiquitin ligases. This current study demonstrated that the lncRNA RMST could suppress the growth and metastasis of LUAD cells through ubiquitination and degradation of SOX9, and this process was mediated by FBW7. These findings revealed the important role of RMST in the growth and metastasis of LUAD and further revealed a novel function of RMST as a mediator for the ubiquitination of SOX9 in LUAD cells.
While RMST has been reported to act as a tumor suppressor in various cancers such as thyroid cancer and head and neck squamous cell carcinomas (18,19), other studies have demonstrated that RMST plays a role as a tumor activator (18). In this current study, RMST expression was downregulated in LUAD tumor tissues compared to normal healthy tissues and high expression of RMST was associated with improved survival in LUAD patients. This seemingly contradictory role of RMST in cancer cells suggests that RMST may exhibit cellular- and tissue-specific functions in a context-dependent manner.
SOX9, which acts as a tumor activator, belongs to the sex determining region Y-related HMG-box (SOX) transcription factor family which includes SOX2, SOX4, SOX6, and SOX9 (20). Many studies have shown that SOX9 is aberrantly expressed in various malignancies and is associated with tumor progression in lung adenocarcinoma, breast cancer, and colorectal cancer (21-23). Ma et al. reported that SOX9 accelerated tumor growth and metastasis, and activated the EMT process in breast cancer (21). This was consistent with our observations in LUAD cells. While Ng et al. previously reported an interaction between RMST and SOX2 (6), the present study documented a direct binding between RMST and SOX9 in LUAD cells. It is unclear whether SOX2 and SOX9 have similar structures that enables them to interact with RMST. Furthermore, the possible interaction between RMST and other members of the SOX family warrants further investigation.
The present study revealed that RMST could interact with the SOX9 protein to promote FBW7-mediated ubiquitination and degradation of SOX9. FBW7, an E3 ubiquitin ligase that mediates ubiquitination and degradation of oncoproteins, usually plays a role as a tumor suppressor (24). Many different proteins have been reported as substrates of FBW7, including SOX9 (5,25). This study documented the novel role of RMST as a scaffold which interacts with SOX9 and FBW7. These results were verified by RNA pull-down assays, as well as RIP and co-IP assays. We speculate that the interaction between SOX9 and RMST may affect the structure of SOX9 such that the ubiquitination site of SOX9 is more readily exposed to FBW7. However, the precise mechanisms by which RMST may influence the structure of SOX9 is unclear and warrants further investigation. In this study, we found that RMST inhibited the occurrence and development of lung cancer through SOX9. In the next study, we will study how to regulate RMST to control lung adenocarcinoma.
Conclusions
In conclusion, this investigation demonstrated a novel lncRNA function whereby RMST binds to SOX9 to mediate the ubiquitination of SOX9 through ubiquitin E3 ligase FBW7. These findings provide new insights into the role of lncRNAs in the progression of LUAD which will contribute to the development of potential strategies for the clinical treatment of LUAD.
Acknowledgments
The authors thank Lei Yao for her assistance in the collection of LUAD samples and the associated pathological studies.
Funding: This work was supported by the Nurture Projects for Basic Research of Shanghai Chest Hospital.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-6052
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-6052
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-6052). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The present study was approved by the Ethics Committee of Zhejiang Provincial People’s Hospital and written informed consent was obtained from all patients. Animal experiments were performed under a project license [No. KS(Y)21220] granted by ethics board of Zhejiang Provincial People’s Hospital, in compliance with the Guide for the Care and Use of Laboratory Animals of Zhejiang Provincial People’s Hospital
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mulvey BB, Olcese U, Cabrera JR, et al. An interactive network of long non-coding RNAs facilitates the Drosophila sex determination decision. Biochim Biophys Acta 2014;1839:773-84. [Crossref] [PubMed]
- Liu S, Zhan N, Gao C, et al. Long noncoding RNA CBR3-AS1 mediates tumorigenesis and radiosensitivity of non-small cell lung cancer through redox and DNA repair by CBR3-AS1 /miR-409-3p/SOD1 axis. Cancer Lett 2022;526:1-11. [Crossref] [PubMed]
- Zhang MX, Zhang LZ, Fu LM, et al. Positive feedback regulation of lncRNA PVT1 and HIF2α contributes to clear cell renal cell carcinoma tumorigenesis and metastasis. Oncogene 2021;40:5639-50. [Crossref] [PubMed]
- Liu C, Peng Z, Li P, et al. lncRNA RMST Suppressed GBM Cell Mitophagy through Enhancing FUS SUMOylation. Mol Ther Nucleic Acids 2020;19:1198-208. [Crossref] [PubMed]
- Wang L, Liu D, Wu X, et al. Long non-coding RNA (LncRNA) RMST in triple-negative breast cancer (TNBC): Expression analysis and biological roles research. J Cell Physiol 2018;233:6603-12. [Crossref] [PubMed]
- Ng SY, Bogu GK, Soh BS, et al. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell 2013;51:349-59. [Crossref] [PubMed]
- Peng WX, Koirala P, Zhang W, et al. lncRNA RMST Enhances DNMT3 Expression through Interaction with HuR. Mol Ther 2020;28:9-18. [Crossref] [PubMed]
- Pei YF, He Y, Hu LZ, et al. The Crosstalk between lncRNA-SNHG7/miRNA-181/cbx7 Modulates Malignant Character in Lung Adenocarcinoma. Am J Pathol 2020;190:1343-54. [Crossref] [PubMed]
- Pei YF, Xu XN, Wang ZF, et al. Methyl-CpG Binding Domain Protein 2 Inhibits the Malignant Characteristic of Lung Adenocarcinoma through the Epigenetic Modulation of 10 to 11 Translocation 1 and miR-200s. Am J Pathol 2019;189:1065-76. [Crossref] [PubMed]
- Pei YF, Yin XM, Liu XQ. TOP2A induces malignant character of pancreatic cancer through activating β-catenin signaling pathway. Biochim Biophys Acta Mol Basis Dis 2018;1864:197-207. [Crossref] [PubMed]
- Pei YF, Lei Y, Liu XQ. MiR-29a promotes cell proliferation and EMT in breast cancer by targeting ten eleven translocation 1. Biochim Biophys Acta 2016;1862:2177-85. [Crossref] [PubMed]
- Jiang Y, Guo H, Tong T, et al. Long noncoding RNA lnc-POP1-1 upregulated by VN1R5 promotes cisplatin resistance in head and neck squamous cell carcinoma through interaction with MCM5. Mol Ther 2022;30:448-67. [Crossref] [PubMed]
- Kawai T, Yasuchika K, Ishii T, et al. SOX9 is a novel cancer stem cell marker surrogated by osteopontin in human hepatocellular carcinoma. Sci Rep 2016;6:30489. [Crossref] [PubMed]
- Panda M, Tripathi SK, Biswal BK. SOX9: An emerging driving factor from cancer progression to drug resistance. Biochim Biophys Acta Rev Cancer 2021;1875:188517. [Crossref] [PubMed]
- Nozaki M, Nishizuka M. Repression of RhoJ expression promotes TGF-β-mediated EMT in human non-small-cell lung cancer A549cells. Biochem Biophys Res Commun 2021;566:94-100. [Crossref] [PubMed]
- Brabletz S, Schuhwerk H, Brabletz T, et al. Dynamic EMT: a multi-tool for tumor progression. EMBO J 2021;40:e108647. [Crossref] [PubMed]
- Lu W, Cao F, Feng L, et al. LncRNA Snhg6 regulates the differentiation of MDSCs by regulating the ubiquitination of EZH2. J Hematol Oncol 2021;14:196. [Crossref] [PubMed]
- Luzón-Toro B, Villalba-Benito L, Fernández RM, et al. RMRP, RMST, FTX and IPW: novel potential long non-coding RNAs in medullary thyroid cancer. Orphanet J Rare Dis 2021;16:4. [Crossref] [PubMed]
- Zhang C, Cao W, Wang J, et al. A prognostic long non-coding RNA-associated competing endogenous RNA network in head and neck squamous cell carcinoma. PeerJ 2020;8:e9701. [Crossref] [PubMed]
- Aksoy I, Jauch R, Eras V, et al. Sox transcription factors require selective interactions with Oct4 and specific transactivation functions to mediate reprogramming. Stem Cells 2013;31:2632-46. [Crossref] [PubMed]
- Ma Y, Shepherd J, Zhao D, et al. SOX9 Is Essential for Triple-Negative Breast Cancer Cell Survival and Metastasis. Mol Cancer Res 2020;18:1825-38. [Crossref] [PubMed]
- Li Y, Zhang X, Zhu S, et al. HDAC10 Regulates Cancer Stem-Like Cell Properties in KRAS-Driven Lung Adenocarcinoma. Cancer Res 2020;80:3265-78. [Crossref] [PubMed]
- Zhou T, Wu L, Ma N, et al. SOX9-activated FARSA-AS1 predetermines cell growth, stemness, and metastasis in colorectal cancer through upregulating FARSA and SOX9. Cell Death Dis 2020;11:1071. [Crossref] [PubMed]
- Xu F, Li J, Ni M, et al. FBW7 suppresses ovarian cancer development by targeting the N6-methyladenosine binding protein YTHDF2. Mol Cancer 2021;20:45. [Crossref] [PubMed]
- Suryo Rahmanto A, Savov V, Brunner A, et al. FBW7 suppression leads to SOX9 stabilization and increased malignancy in medulloblastoma. EMBO J 2016;35:2192-212. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


