
A transcriptomic analysis based on aberrant methylation levels revealed potential novel therapeutic targets for nasopharyngeal carcinoma
Introduction
Nasopharyngeal carcinoma (NPC) is a kind of epithelial malignant tumor, which is the most common type of otolaryngological cancer. According to statistics of the World Health Organization, NPC has obvious geographical distribution characteristics and an ethnic tendency (1). The incidence of NPC is very low on Africa, Europe, and America. However, the incidence of NPC is significantly increased in China and Southeast Asia (2). Indeed, China has the highest incidence of NPC worldwide (2). The rate of nasopharyngeal cancer in Guangdong, which has a high incidence of NPC is dozens times higher than the average rate of NPC in the world. Thus, NPC is also called “Guangdong cancer” (3).
NPC is prone to local invasion and distant metastasis, and has a poor prognosis (4). As the cancer occurs in the nasopharynx, and is constrained by its anatomical structure and physiological characteristics, radiotherapy is the first choice for the treatment of early lesions (5). In advanced and recurrent patients, as the sensitivity of cancer cells to radiotherapy is reduced after radiotherapy, chemotherapy and surgical resection are the preferred treatment strategies (5).
NPC is induced by multiple factors. Epstein-Barr virus (EBV) infection, heritage susceptibility, environmental carcinogens, and eating habits are potential reasons for the occurrence and development of NPC (6-8). EBV proliferates massively in the oropharyngeal region and infects the whole body through lymphocytes, and is closely related to the incidence of NPC (6). Additionally, NPC cells are often genetically susceptible to changes on chromosomes 1, 3, 11, 12, and 17 (7). The people of Guangdong, China, like to eat salted fish and other products rich in nitrites, which are a potential cause of nasopharyngeal cancer (8).
The occurrence of cancer is accompanied by a series of complex molecular changes in cells. DNA methylation is a common epigenetic modification, most of which occurs at the fifth carbon atom of cytosine-phosphate-guanine in a specific gene region (9). It is involved in important biological processes such as gene expression, embryonic development, gene imprinting and X chromosome inactivation. In addition, it affects cell susceptibility, tumor phenotype, and tumor malignancy (10). Studies have shown that plasma EBV DNA methylation map of patients with nasopharyngeal carcinoma has important potential for the diagnosis of nasopharyngeal carcinoma (11). SHISA3 hypermethylation silences the gene expression and leads to poor prognosis of nasopharyngeal carcinoma (12), while ARNTL hypermethylation leads to significant down-regulation of its mRNA and protein in nasopharyngeal carcinoma cell lines and tissues (13). These studies show that DNA methylation patterns and abnormal gene expression often occur in nasopharyngeal carcinoma, aberrant methylation and differentially expressed genes can be used as potential biomarkers for cancer treatment and prediction. Nearly 80% of methylation levels were negatively correlated with gene expression levels, and a few were positively correlated (14). It was generally consistent with the view that the increase of methylation level in tumors was related to transcriptional silencing.
With the continuous development of modern omics technology, and bioinformatics, a large number of studies on the pathogenesis of NPC have emerged, and scholars at home and abroad have reported relevant research results one after another (15). The pathogenesis of NPC and the mechanism of drug resistance have been explained at the molecular level (16,17). In this study, we integrated NPC gene expression and methylation data sets published on the Gene Expression Omnibus (GEO) database. Under the search terms set in this study, the most comprehensive gene expression and methylation data available for NPC were included. Integrating multiple data sets from multiple experiments not only increases the sample size, but also improves the robustness of the results, which is of great significance to the in-depth exploration of the molecular changes in NPC. The identification of important expression pathways and potential oncogenes in NPC will provide new directions and ideas for clinicians, and assist in the identification of diagnostic markers and therapeutic targets.
We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6628/rc).
Methods
Data sources, filtering and selection
The flow chart of the research process is shown in Figure 1. Human microarray and high-throughput sequencing gene expression NPC data sets were retrieved from the National Center for Biotechnology Information’s GEO database (http://www.ncbi.nlm.nih.gov/geo/) by searching for “Nasopharyngeal carcinoma”, “Series”, “Homo sapiens”, “Expression profiling by array”, “Expression profiling by high-throughput sequencing”, and “Methylation profiling by array”. The GEO data sets were then filtered according to the criteria below: (I) the gene expression profile had been exclusively derived from the human nasopharyngeal tissue of patient and healthy control groups and probed using a human-based genome array platform; (II) experiments containing long non-coding ribonucleic acid (RNA), microRNA, small RNA, and single-cell RNA sequencing were excluded from this study; (III) each study and control group had at least 3 samples. After filtering based on the above-mentioned conditions, 10 data sets (i.e., GSE12452, GSE13597, GSE40290, GSE53819, GSE64634, GSE118719, GSE68799, GSE134886, GSE52068, and GSE62336) of human nasopharyngeal tissue were obtained using the getGEO function of the GeoQuery package in R software (version 3.6.1) for further analysis. All the retrieved data sets were microarray data sets except for GSE118719, GSE68799, and GSE134886, which were high-throughput sequencing gene expression data sets. GSE12452, GSE13597, GSE40290, GSE53819, GSE64634, GSE118719, GSE68799, and GSE134886 were gene expression data sets, while GSE52068 and GSE62336 were deoxyribonucleic acid (DNA) methylation data sets. Raw gene expression data, study design tables, and annotation tables of each data set were obtained from the GEO database. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
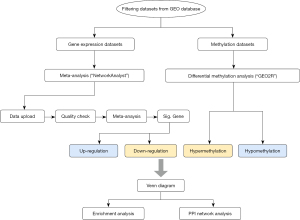
Data processing and meta-analysis of the gene expression data sets
NetworkAnalyst (https://www.networkanalyst.ca/) is an online tool for analyzing gene expression data (18-21). Eight gene expression data sets were uploaded to the “Multiple Gene Expression Tables” module on the homepage of the website for the meta-analysis. All probe/gene IDs were first converted into the Entrez ID format. The NetworkAnalyst online site has a library of 47 human, mouse, and rat on-chip data probe identifiers (IDs). The probe platforms for the 8 gene expression data sets were Affymetrix Human Genome U133plus2 (HGU133plus2), Affymetrix Human Genome U133 (chip hgu133a), Ensemble Transcript ID, Agilent Human Genome Whole Microarray (4x44k/4112), Affymetrix Human Genome U133plus2 (HGU133plus2), Official Gene Symbol, Ensembl Gene ID, and Ensembl Gene ID. Variance stabilization normalization and log2 counts per million methods were adopted to standardize the data sets. The Limma method was used to analyze the differential expression of the 8 gene expression data sets. The cut-off P values were adjusted using Benjamini-Hochberg’s rate of error discovery [i.e., the false discovery rate (FDR)]. A P value of 0.05 was considered statistically significant. After the data preparation was completed, a total of 5,208 features were matched with a total sample size of 217. The groups were divided into “Ctrl” and “NPC” groups. The combat method was used to adjust the batch effects of the data. A principal component analysis was conducted, and density plots of the gene counts were generated, using the merged data. The meta-analysis was conducted after a data quality check. A random model of combining effect sizes was used to combine data sets from multiple studies. The significance level was set to 0.05. For the 2 DNA methylation data sets (GSE52068 and GSE62336), we used GEO2R to identify differential methylation sites. GEO2R performs comparisons on original submitter-supplied processed data tables using the GEOquery and limma R packages from the Bioconductor project. The significance level was set to 0.05.
Aberrantly methylated DEGs
Studies have shown that the pathogenesis of NPC is often accompanied by methylation variation, and DNA methylation often inhibits gene expression (22). We took the intersections of the downregulated genes from the meta-analysis and the hypermethylated genes from GEO2R and drew a Venn map on the “Bioinformatics & Evolutionary Genomics” website (http://bioinformatics.psb.ugent.be/webtools/Venn/). Similarly, we also took the intersections of upregulated genes from the meta-analysis and the hypomethylated genes from GEO2R and drew a Venn map on the same website mentioned above.
Enrichment analysis
A functional enrichment analysis was performed for the aberrantly methylated differentially expressed genes (DEGs) on the website “Metascape” (23). For these genes, the pathway and process enrichment analysis was performed using the following ontology sources: Gene Ontology (GO) Biological Processes, GO Cellular Components, GO Molecular Functions, the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway, and Reactome Gene Sets. The criteria for selecting the items were: (I) P value is less than 0.01; (II) the minimum count threshold is 3; (III) the minimum enrichment factor threshold is 1.5. In order to further capture the relationship between terms, we select a subset of enriched terms and present it as a network graph, in which terms with similarity index greater than 0.3 are connected by edges. We selected the terms with the best P value from 20 clusters, and restrict each cluster to no more than 15 terms, with a total of no more than 250 terms.
Protein-protein interaction (PPI) network analysis
The “String” website was used to analyze the PPI network of each of the aberrantly methylated DEGs (24). The active interaction sources included text mining, experiments, databases, co-expression, neighborhood, gene fusion, and co-occurrence. The minimum required interaction score was a medium confidence level of 0.400, and unrelated gene nodes in the network were hidden. The preliminary network results were obtained and exported to -tab separated values format for further analysis. The local PPI analysis software “Cytoscape” was then used to retouch the network and for further analysis (25,26). “MCODE” and “cytohubba” modules were carried out to identify the key network nodes and hub genes, for which the degree cut-off value was set as 2, the node cut-off score was set as 0.2, the k-core was set as 2, the max depth was set as 100, and the Maximal Clique Centrality (MCC) algorithm was used.
Statistical analysis
Most of the statistical methods used in this study were based on the above-mentioned bioinformatics online or local analysis tools. The Limma package was used to identify the DEGs. Benjamin Hochberg’s FDR was used to adjust the cut-off P value, and to combine effect sizes, a random-effects model was used to combine the data sets from multiple studies. In all the statistical analyses, P value less than 0.05 was considered statistically significant
Results
Data information included in this study
Ten data sets (i.e., GSE12452, GSE13597, GSE40290, GSE53819, GSE64634, GSE118719, GSE68799, GSE134886, GSE52068, and GSE62336) from human nasopharyngeal tissue were obtained for further analysis after filtering in accordance with the conditions mentioned in the “Methods” section above. In total, 315 samples were included in our study. The study type, accession number, PubMed Unique Identifier (PMID), Platform, sample source, and sample size of these data sets are set out in Table 1.
Table 1
| Study type | Accession | PMID | Platform | Source | Samples | |
|---|---|---|---|---|---|---|
| NPC | Ctrl | |||||
| Expression profiling by array | GSE12452 | 17119049; 16912175; 22880099 | Affymetrix | Nasopharyngeal tissue | 31 | 10 |
| Expression profiling by array | GSE13597 | 19142888 | Affymetrix | Nasopharyngeal tissue | 25 | 3 |
| Expression profiling by array | GSE40290 | NA | Capitalbio | Nasopharyngeal tissue | 25 | 8 |
| Expression profiling by array | GSE53819 | 24763226 | Agilent | Nasopharyngeal tissue | 18 | 18 |
| Expression profiling by array | GSE64634 | 26246469 | Affymetrix | Nasopharyngeal tissue | 12 | 4 |
| Expression profiling by high-throughput sequencing | GSE118719 | 30477559 | Illumina | Nasopharyngeal tissue | 7 | 4 |
| Expression profiling by high-throughput sequencing | GSE68799 | NA | Illumina | Nasopharyngeal tissue | 42 | 4 |
| Expression profiling by high-throughput sequencing | GSE134886 | 33931030 | HiSeq | Nasopharyngeal tissue | 3 | 3 |
| Methylation profiling by array | GSE52068 | 26443805; 28146149 | Illumina | Nasopharyngeal tissue | 24 | 24 |
| Methylation profiling by array | GSE62336 | 25924914 | Illumina | Nasopharyngeal tissue | 25 | 25 |
| Total | 212 | 103 | ||||
GEO, Gene Expression Omnibus.
Gene expression data processing and meta-analysis
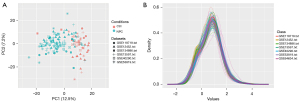
To check the distribution of the data and whether there were abnormal values before the enrichment and PPI network analyses, we used a 2-dimensional principal component analysis and density plots of the gene counts to check the data, and the results showed that there was good differentiation between the NPC group and the normal control group (see Figure 2).
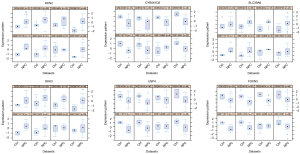
First, in relation to the meta-analysis, 8 data sets were analyzed by DEGs, respectively. The numbers of the GSE12452, GSE13597, GSE40290, GSE53819, GSE64634, GSE118719, GSE68799, and GSE134886 DEG data sets were 4,968, 1, 640, 6,227, 99, 7,968, 2,617, and 0, respectively. Second, the “Multiple Gene Expression Tables” of the “NetworkAnalyst” were used to integrate and analyze the 8 groups of data. In total, 1,488 DEGs were identified by combining the effect sizes using the random-effects model method. 721 genes were downregulated and 767 genes were upregulated. The top 50 genes with P values are set out in Table 2. The boxplot of representative differential genes is shown in Figure 3, and all the genes are specified in https://cdn.amegroups.cn/static/public/atm-21-6628-1.xlsx.
Table 2
| EntrezID | Name | CombinedES | Pval |
|---|---|---|---|
| 5955 | RCN2 | 1.6706 | 1.35E-09 |
| 11068 | CYB561D2 | –1.2929 | 2.06E-09 |
| 25800 | SLC39A6 | 1.3176 | 2.35E-09 |
| 27122 | DKK3 | 1.2715 | 2.35E-09 |
| 7375 | USP4 | –1.2787 | 5.14E-09 |
| 1112 | FOXN3 | –1.5158 | 5.26E-09 |
| 25888 | ZNF473 | 1.2064 | 1.81E-08 |
| 1054 | CEBPG | 1.1895 | 2.19E-08 |
| 10947 | AP3M2 | 1.176 | 3.72E-08 |
| 2288 | FKBP4 | 1.3345 | 6.40E-08 |
| 22911 | WDR47 | 1.1418 | 7.89E-08 |
| 4350 | MPG | –1.1404 | 7.89E-08 |
| 10651 | MTX2 | 1.136 | 8.91E-08 |
| 6591 | SNAI2 | 1.1299 | 8.91E-08 |
| 9208 | LRRFIP1 | –1.1225 | 1.48E-07 |
| 9378 | NRXN1 | 1.3541 | 1.57E-07 |
| 53616 | ADAM22 | 1.3295 | 1.68E-07 |
| 11060 | WWP2 | –1.2219 | 1.81E-07 |
| 931 | MS4A1 | –1.5446 | 1.81E-07 |
| 9450 | LY86 | –1.4976 | 2.52E-07 |
| 6039 | RNASE6 | –1.1407 | 2.52E-07 |
| 5525 | PPP2R5A | –1.416 | 3.99E-07 |
| 3685 | ITGAV | 1.9058 | 3.99E-07 |
| 23345 | SYNE1 | –1.3434 | 3.99E-07 |
| 9258 | MFHAS1 | 1.057 | 3.99E-07 |
| 4430 | MYO1B | 1.4312 | 3.99E-07 |
| 7351 | UCP2 | –1.3914 | 4.25E-07 |
| 6770 | STAR | 1.351 | 5.81E-07 |
| 1462 | VCAN | 1.2441 | 5.81E-07 |
| 25864 | ABHD14A | –1.3261 | 6.04E-07 |
| 933 | CD22 | –1.8081 | 6.21E-07 |
| 23231 | SEL1L3 | –1.1697 | 7.54E-07 |
| 1535 | CYBA | –1.0378 | 7.79E-07 |
| 23550 | PSD4 | –1.4916 | 7.95E-07 |
| 7130 | TNFAIP6 | 1.8405 | 8.52E-07 |
| 640 | BLK | –1.9199 | 8.52E-07 |
| 8508 | NIPSNAP1 | 1.0292 | 8.87E-07 |
| 4232 | MEST | 1.0322 | 9.96E-07 |
| 930 | CD19 | –1.5597 | 1.02E-06 |
| 2690 | GHR | 1.0108 | 1.32E-06 |
| 54677 | CROT | 1.0081 | 1.40E-06 |
| 10520 | ZNF211 | –1.008 | 1.71E-06 |
| 55884 | WSB2 | 0.99518 | 1.98E-06 |
| 8050 | PDHX | 0.99695 | 2.07E-06 |
| 7480 | WNT10B | 0.99621 | 2.08E-06 |
| 4902 | NRTN | 0.99854 | 2.19E-06 |
| 81628 | TSC22D4 | –0.99859 | 2.26E-06 |
| 23221 | RHOBTB2 | –1.1857 | 2.37E-06 |
| 8850 | KAT2B | –0.98947 | 2.37E-06 |
| 191 | AHCY | 1.4157 | 2.51E-06 |
DEGs, differentially expressed genes.
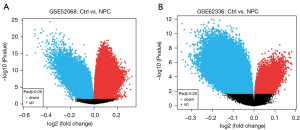
Aberrantly methylated gene loci
A total of 21,717 differentially methylated loci were identified in the GSE62336 data set, of which 5,800 were hypermethylated and 15,917 were hypomethylated. Additionally, a total of 30,426 differentially methylated loci were identified in the GSE52068 data set, of which 17,128 were hypermethylated and 13,298 were hypomethylated. The volcano map of differentially methylated genes is shown in Figure 4.
Aberrantly methylated DEGs
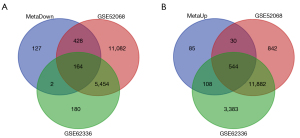
The Venn diagram analysis integrated the DEGs from the meta-analysis and the differential methylation genes. In practical terms, a total of 164 hypermethylation low-expression genes were obtained by overlapping the hypermethylation genes from GSE52068, GSE62336, and the 721 downregulated genes from the meta-analysis, and 544 hypomethylation high-expression genes were obtained by overlapping the hypomethylation genes from GSE52068, GSE62336, and the 767 upregulated genes from the meta-analysis. The Venn diagrams are shown in Figure 5.
Enrichment pathway and process
The enrichment results showed that the 164 hypermethylation low-expression genes were enriched in the biological processes of cellular response to nitrogen compounds, regulation of ion transmembrane transport, actin filament-based process, response to drugs, positive regulation of cell death, myelination, cellular calcium ion homeostasis, and regulation of small GTPase-mediated signal transduction. In relation to the cellular components, the genes were mainly enriched in the extrinsic components of the plasma membrane, sarcomere, and perinuclear region of cytoplasm. In relation to molecular function, the genes were mainly enriched in enzyme activator activity and hormone receptor binding. In relation to the KEGG pathway, the genes were mainly enriched in the calcium signaling pathway, the mitogen-activated protein kinase signaling pathway, and inositol phosphate metabolism. Finally, the genes in the Reactome Gene Sets were mainly enriched in G protein-coupled receptor (GPCR) downstream signaling, transmission across chemical synapses, neural cell adhesion molecule (NCAM) signaling for neurite out-growth, and PI5P, PP2A and IER3 regulate PI3K/AKT signaling (see Figure 6A).
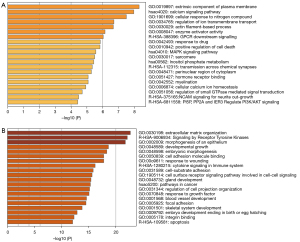
The enrichment results showed that the 544 hypomethylation high-expression genes were enriched in the biological processes of extracellular matrix organization, epithelium morphogenesis, developmental growth, embryonic morphogenesis, response to wounding, cell-substrate adhesion, the cell surface receptor signaling pathway involved in cell-cell signaling, gland development, the regulation of cell projection organization, growth-factor responses, blood vessel development, skeletal system development, and embryo development ending in birth or egg hatching. In relation to the cellular components, the genes were mainly enriched in focal adhesion. In relation to molecular function, the genes were mainly enriched in cell adhesion molecule binding and integrin binding. In relation to the KEGG pathway, the genes were mainly enriched in pathways in cancer. Finally, the genes in the Reactome Gene Sets were mainly enriched in signaling by receptor tyrosine kinases, cytokine signaling in the immune system, and apoptosis (see Figure 6B).
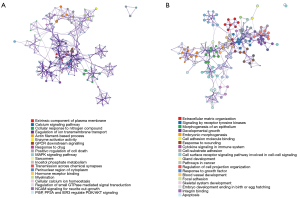
To further pick out modules that are momentous, accumulative hypergeometric p values and enrichment factors of all the statistically significant enriched terms were calculated. Then, the remaining significant terms were hierarchically clustered into a tree according to the kappa statistical similarities among their gene members, The tree convert into term clusters according to the Kappa score threshold of 0.3. And select a subset of representative terms from the cluster to present the network graph. Figure 7 shows the network plots that capture the relationships between the terms. Each term is represented by a circular node, the larger the node, the more genes the term has entered, and the colors of the node indicate different clusters. Terms with a similarity score higher than 0.3 are connected by an edge. The thicker the edges, the higher the similarity score.
PPI network construction
The two groups of aberrantly methylated DEGs were imported into the “String” website, and 164/544 nodes were correctly identified in the network, with a total of 187/3,134 pairs of interactions. The average node level was 2.28/11.5. The average local clustering coefficient was 0.283/0.35. The expected number of edges was 130/2,294. The P value of PPI enrichment was 1.59e-06/1.0e-16. After hiding the unrelated gene nodes in the network, the results were imported into “Sytoscape,” and the “MCODE” plug-in was used to identify more meaningful modules in the network. The parameters of the mcode were set to the default parameters. The top 3 modules of hypermethylation downregulated genes are illustrated in Figure 8A. The enrichment analysis revealed that their main functions related to the T cell receptor signaling pathway, signal transduction, and carbon metabolism (see Table 3). The top 10 hub genes for hypermethylation low-expression were ITPKB, GNB5, FYN, LCK, NFATC1, GNAS, PRKCB, ZAP70, LPAR1, and PRKCE (see Figure 8B). In relation to the PPI network of hypomethylation upregulated genes, the functions of the top 3 modules were related to the regulation of protein metabolic process, cancer pathways, and protein-containing complexes (see Figure 8C and Table 3). The top 10 hub genes for hypomethylation high expression were TP53, GAPDH, FN1, CCND1, VEGFA, HRAS, STAT3, FGF2, APP, and MMP2 (see Figure 8D).
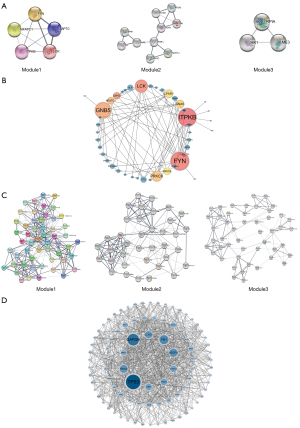
Table 3
| Group | Module | Category | Description | Genes | Nodes | FDR |
|---|---|---|---|---|---|---|
| Hyper-low | 1 | KEGG pathways | T-cell receptor signaling pathway | ZAP70, LCK, FYN, NFATC1 | 4 | 0.00000013 |
| 2 | Reactome pathways | Signal transduction | PTH, PNOC, TACR1, P2RY1, VIPR1, LPAR1 | 6 | 0.0000745 | |
| 3 | KEGG pathways | Carbon metabolism | RPIA, HK1, ME3 | 3 | 0.00000351 | |
| Hypo-high | 1 | GO process | Regulation of protein metabolic process | TNFRSF1A, KEAP1, CCND1, GAPDH, RNF114, CDK4… | 36 | 0.000000000000000683 |
| 2 | KEGG pathways | Pathways in cancer | LAMB1, KITLG, JAG1, LAMC1, CCND2, LRP6, FZD7… | 17 | 0.00000000000000132 | |
| 3 | GO component | Protein-containing complexes | DLD, COX6A1, ENO1, LRP1, CANX, DDB2, COX5B, NUP133… | 36 | 0.0000000001 |
PPI, protein-protein interaction.
Discussion
The molecular mechanisms underlying the development and progression of and potential novel therapeutic targets for NPC need to be explored. DNA methylation is a form of DNA chemical modification that can change the genetic performance without changing the DNA sequence. A large number of studies have shown that DNA methylation plays an important role in chromatin structure, DNA conformation, DNA stability, and the interaction between DNA and protein, thus controlling the expression of target genes (27). Studies have also indicated that the pathological changes of NPC are often accompanied by methylation changes (28). With that in mind, in this study, based on the previous rich research results on the molecular level of NPC and the increasingly in-depth research with sequencing technology in recent years, we analyzed NPC gene expression and DNA methylation data sets from 10 groups of experiments. Aberrantly methylated DEGs and pathways were obtained for further enrichment and PPI network analyses.
For the enrichment analysis, we focused on the enrichment results from the Reactome Gene Sets, which had more detailed and specific significance than other enrichment databases. In relation to the 164 hypermethylation low-expression genes, the enrichment results showed that these genes were enriched in GPCR downstream signaling, transmission across chemical synapses, and NCAM signaling for neurite out-growth. This is reasonable, as studies have shown that the downstream regulators of GPCRs, such as the Hippo signaling pathway, are involved in the regulation of tumor size and tumorigenesis (29). As for transmission across chemical synapses and NCAM signaling for neurite out-growth, there is no literature on the relationship between these two pathways and cancer, especially NPC. In relation to the 544 hypomethylation high-expression genes, the enrichment results showed that these genes were enriched in signaling by receptor tyrosine kinases, cytokine signaling in the immune system, and apoptosis. These results are credible, as receptor tyrosine kinases (RTKs) have been shown to play an important role in cell growth, movement, differentiation, and metabolism (30). Thus, an imbalance in RTK signaling leads to a variety of human diseases, including cancer (31). Additionally, cytokines are one of the most important effector and messenger molecules in the immune system. They are deeply involved in immune responses during infection and inflammation, and in the prevention or promotion of diseases, such as allergies and cancer. Thus, regulating the cytokine pathway is one of the most effective strategies for treating various diseases (32). It has also been reported that the signaling of tyrosine kinase is related to the radiation resistance of NPC (33). Many studies have shown that the apoptosis of NPC cells can be induced through the regulation of target genes (34,35).
Next, the PPI network analysis examined the relationship among proteins encoded by these aberrantly methylated DEGs. The top 10 hub genes in the hypermethylation low-expression group were ITPKB, GNB5, FYN, LCK, NFATC1, GNAS, PRKCB, ZAP70, LPAR1, and PRKCE. The presence of ITPKB has been reported in lung cancer metastasis and acute and chronic graft-versus-host diseases, and has been used in tumor therapy interventions to overcome cisplatin resistance (36-38). ITPKB has not been examined in NPC, and it may be a potential novel target. FYN is a non-receptor tyrosine kinase, belonging to the sarcoma (Src) family of kinases. It is involved in the signal transduction pathways of the nervous system and in the development and activation of T lymphocytes under normal physiological conditions. FYN has been shown to contribute to the development and progression of several types of cancer by participating in the control of cell growth, death, morphogenesis, and cell metastasis (39). In addition, the top 10 hub genes in the hypomethylation high-expression group were TP53, GAPDH, FN1, CCND1, VEGFA, HRAS, STAT3, FGF2, APP, and MMP2. TP53, the star gene in cancer, also appeared in our results, which plays an important role in many cancers. GAPDH is a multifunctional enzyme, which plays a variety of regulatory roles in determining cell fate (40). To date, very little research has been conducted on the regulatory mechanism of GAPDH in NPC.
Regarding the relationship between the prognosis of nasopharyngeal carcinoma and methylation, studies have shown that nasopharyngeal carcinoma with high methylation levels is significantly associated with poorer survival (41). This is consistent with the fact that hypermethylation leads to low expression of tumor suppressor genes and hypomethylation leads to high expression of oncogenes.
In conclusion, our combined bioinformatics analysis of gene expression and gene methylation microarrays revealed a series of DEGs and pathways of abnormal methylation in NPC. The results may help to reveal the molecular mechanism underling the occurrence and development of NPC and provide new ideas for the targeted therapy of NPC. Hub genes, including ITPKB, GNB5, FYN, LCK, NFATC1, GNAS, PRKCB, ZAP70, LPAR1, PRKCEW, TP53, GAPDH, FN1, CCND1, VEGFA, HRAS, STAT3, FGF2, APP, and MMP2, are noteworthy. Under the search terms set in this study, the most comprehensive gene expression and methylation data available for NPC were included, and this study produced more reliable and accurate screening results than individual surveys by overlapping relevant data sets. A number of reliable online and local analysis software was used to identify pathways and key sites that might have been overlooked in previous studies. However, this study had some limitations. For example, due to the variable data information, we could not obtain all the clinical information of the data, and the effects of the Hub genes on prognosis and metastasis still needs to be further studied and verified.
Conclusions
In our data analysis, we identified novel important pathways and genes pivotal to the development of NPC. Our findings can guide clinical research and could lead to the development of drug targets that act on relevant pathways and genes.
Acknowledgments
Funding: This research was supported by grants from the National Natural Science Foundation of China (No. 82171931), Natural Science Foundation of Guangdong (No. 2015A030313753), the Science and Technology Program of Guangzhou (Nos. 201903010032 and 202102080572), and the Panyu Science and Technology Program of Guangzhou (Nos. 2017-Z04-12, 2019-Z04-01, and 2019-Z04-23).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6628/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6628/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Chen YP, Chan ATC, Le QT, et al. Nasopharyngeal carcinoma. Lancet 2019;394:64-80. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Ren X, Yang X, Cheng B, et al. HOPX hypermethylation promotes metastasis via activating SNAIL transcription in nasopharyngeal carcinoma. Nat Commun 2017;8:14053. [Crossref] [PubMed]
- Chan AT, Grégoire V, Lefebvre JL, et al. Nasopharyngeal cancer: EHNS-ESMO-ESTRO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2012;23:vii83-5. [Crossref] [PubMed]
- Tsao SW, Tsang CM, Lo KW. Epstein-Barr virus infection and nasopharyngeal carcinoma. Philos Trans R Soc Lond B Biol Sci 2017;372:20160270. [Crossref] [PubMed]
- Huang DP, Ho JH, Chan WK, et al. Cytogenetics of undifferentiated nasopharyngeal carcinoma xenografts from southern Chinese. Int J Cancer 1989;43:936-9. [Crossref] [PubMed]
- Ward MH, Pan WH, Cheng YJ, et al. Dietary exposure to nitrite and nitrosamines and risk of nasopharyngeal carcinoma in Taiwan. Int J Cancer 2000;86:603-9. [Crossref] [PubMed]
- Kulis M, Esteller M. DNA methylation and cancer. Adv Genet 2010;70:27-56. [Crossref] [PubMed]
- Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet 2018;392:777-86. [Crossref] [PubMed]
- Lam WKJ, Jiang P, Chan KCA, et al. Methylation analysis of plasma DNA informs etiologies of Epstein-Barr virus-associated diseases. Nat Commun 2019;10:3256. [Crossref] [PubMed]
- Zhang J, Li YQ, Guo R, et al. Hypermethylation of SHISA3 Promotes Nasopharyngeal Carcinoma Metastasis by Reducing SGSM1 Stability. Cancer Res 2019;79:747-59. [Crossref] [PubMed]
- Peng H, Zhang J, Zhang PP, et al. ARNTL hypermethylation promotes tumorigenesis and inhibits cisplatin sensitivity by activating CDK5 transcription in nasopharyngeal carcinoma. J Exp Clin Cancer Res 2019;38:11. [Crossref] [PubMed]
- Zouridis H, Deng N, Ivanova T, et al. Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci Transl Med 2012;4:156ra140. [Crossref] [PubMed]
- Janvilisri T. Omics-based identification of biomarkers for nasopharyngeal carcinoma. Dis Markers 2015;2015:762128. [Crossref] [PubMed]
- Paul P, Deka H, Malakar AK, et al. Nasopharyngeal carcinoma: understanding its molecular biology at a fine scale. Eur J Cancer Prev 2018;27:33-41. [Crossref] [PubMed]
- Chou J, Lin YC, Kim J, et al. Nasopharyngeal carcinoma--review of the molecular mechanisms of tumorigenesis. Head Neck 2008;30:946-63. [Crossref] [PubMed]
- Zhou G, Soufan O, Ewald J, et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res 2019;47:W234-41. [Crossref] [PubMed]
- Xia J, Gill EE, Hancock RE. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc 2015;10:823-44. [Crossref] [PubMed]
- Xia J, Lyle NH, Mayer ML, et al. INVEX--a web-based tool for integrative visualization of expression data. Bioinformatics 2013;29:3232-4. [Crossref] [PubMed]
- Xia J, Fjell CD, Mayer ML, et al. INMEX--a web-based tool for integrative meta-analysis of expression data. Nucleic Acids Res 2013;41:W63-70. [Crossref] [PubMed]
- Han B, Yang X, Zhang P, et al. DNA methylation biomarkers for nasopharyngeal carcinoma. PLoS One 2020;15:e0230524. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Otasek D, Morris JH, Bouças J, et al. Cytoscape Automation: empowering workflow-based network analysis. Genome Biol 2019;20:185. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Varizhuk A, Isaakova E, Pozmogova G. DNA G-Quadruplexes (G4s) Modulate Epigenetic (Re)Programming and Chromatin Remodeling: Transient Genomic G4s Assist in the Establishment and Maintenance of Epigenetic Marks, While Persistent G4s May Erase Epigenetic Marks. Bioessays 2019;41:e1900091. [Crossref] [PubMed]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell 2012;150:12-27. [Crossref] [PubMed]
- Luo J, Yu FX. GPCR-Hippo Signaling in Cancer. Cells 2019;8:426. [Crossref] [PubMed]
- Shi Q, Chen YG. Interplay between TGF-β signaling and receptor tyrosine kinases in tumor development. Sci China Life Sci 2017;60:1133-41. [Crossref] [PubMed]
- Du Z, Lovly CM. Mechanisms of receptor tyrosine kinase activation in cancer. Mol Cancer 2018;17:58. [Crossref] [PubMed]
- Ouyang W, O'Garra A. IL-10 Family Cytokines IL-10 and IL-22: from Basic Science to Clinical Translation. Immunity 2019;50:871-91. [Crossref] [PubMed]
- Shen L, Li Z, Shen L. Quantitative Tyrosine Phosphoproteomic Analysis of Resistance to Radiotherapy in Nasopharyngeal Carcinoma Cells. Cancer Manag Res 2020;12:12667-78. [Crossref] [PubMed]
- Sun FR, Wang SL, Wang M, et al. Simvastatin induces apoptosis of nasopharyngeal carcinoma cells through NF-κB signaling pathway. Eur Rev Med Pharmacol Sci 2020;24:6726-34. [PubMed]
- Wang TT, Chen ZZ, Xie P, et al. Isoliquiritigenin suppresses the proliferation and induced apoptosis via miR-32/LATS2/Wnt in nasopharyngeal carcinoma. Eur J Pharmacol 2019;856:172352. [Crossref] [PubMed]
- Bäder S, Glaubke E, Grüb S, et al. Effect of the actin- and calcium-regulating activities of ITPKB on the metastatic potential of lung cancer cells. Biochem J 2018;475:2057-71. [Crossref] [PubMed]
- Pan C, Jin L, Wang X, et al. Inositol-triphosphate 3-kinase B confers cisplatin resistance by regulating NOX4-dependent redox balance. J Clin Invest 2019;129:2431-45. [Crossref] [PubMed]
- Thangavelu G, Du J, Paz KG, et al. Inhibition of inositol kinase B controls acute and chronic graft-versus-host disease. Blood 2020;135:28-40. [Crossref] [PubMed]
- Elias D, Ditzel HJ. Fyn is an important molecule in cancer pathogenesis and drug resistance. Pharmacol Res 2015;100:250-4. [Crossref] [PubMed]
- Tristan C, Shahani N, Sedlak TW, et al. The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal 2011;23:317-23. [Crossref] [PubMed]
- Jiang W, Liu N, Chen XZ, et al. Genome-wide identification of a methylation gene panel as a prognostic biomarker in nasopharyngeal carcinoma. Mol Cancer Ther 2015;14:2864-73. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


