
Implementation of a pathological diagnosis and treatment pathway may improve the molecular detection of lung cancer
Introduction
With the widespread application of next generation sequencing (NGS) in clinical work, a large number of genetic aberrations have been identified, thus permitting the development of new targeted therapies for patients with cancer, including advanced non-small cell lung cancer (NSCLC) (1). Over recent years, new targeted drugs, molecular markers, and detection methods have been constantly emerging (2). The recent treatment guidelines of the National Comprehensive Cancer Network (NCCN) in the United States have suggested that 9 driver genes (EGFR/KRAS/HER2/ALK/ROS1/MET/BRAF/RET/NTRK) should be routinely detected in lung cancer to provide more detailed genetic mutation information for the precise treatment of lung cancer (3).
As an advanced genetic testing method, NGS has the advantages of high sensitivity and comprehensive detection. Compared to traditional methods such as fluorescence in situ hybridization (FISH), immunohistochemistry (IHC), and reverse transcription-polymerase chain reaction (RT-PCR), NGS has a longer detection cycle (results are usually obtained within 4–10 natural days). Recent guideline recommendations by the Association for Molecular Pathology (AMP), the College of American Pathologists (CAP), and the International Association for the Study of Lung Cancer (IASLC) established a benchmark turnaround time (TAT). Based on the guideline, results should be available to the treating oncologist within 10 working days (4,5). In a retrospective analysis of all NSCLC cases from a large academic institution within 1 year, DiStasio et al. found that 81.5% of the test results were received within 10 working days as recommended by the guidelines, which demonstrated the feasibility of the TAT recommended by the CAP, IASLC, and AMP guidelines (6).
In our clinical work, the puncture biopsy specimens are relatively small and may not be sufficient for gene testing after routine HE and immunohistochemistry. This will delay the follow-up treatment of patients, and patients may also face the risk of second puncture biopsy. Therefore, we decided to design a sample collector, “biology collector (BIOCO)” tool, to collect the remaining paraffin samples as much as possible to meet the quality control requirements of gene testing.
In the process of clinical diagnosis and treatment, clinicians often face two problems: firstly, there are not enough puncture biopsy samples; secondly, the time of Next Generation Sequencing (NGS) testing is too long. Therefore, our team established a rapid and accurate diagnosis and genotyping workflow. The clinicians put forward applications for routine pathological diagnosis and genetic testing at the same time, communicated with patients and signed informed consent forms. After the biopsy samples were embedded, “BIOCO” tool was used to collect specimens left over from pruning wax blocks during HE and immunohistochemical detection. What’s more, the largest area of white slices were cut. Gene testing was carried out at the same time with pathological diagnosis. We can obtain enough tissue samples as far as possible and carry out NGS detection on the premise of ensuring pathological diagnosis.
With the development of precision medicine, both patients and clinicians expect to benefit from precise and timely treatment, so a rapid and accurate diagnosis and genotyping workflow for lung cancer are required. In this study, we established and validated a new protocol for a pathological diagnosis and treatment pathway that was tested in our department. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6230/rc).
Methods
Specimens
From January 1, 2019 to November 30, 2019, 384 specimens were collected and detected on the hospital platform, all of which were puncture biopsy specimens. All patients were diagnosed with NSCLC, it’s need to obtain the enough paraffin samples from the same patient to take the Routine Collection (ROUCO) method and BIOCO method. After randomization, 39 specimens were obtained for subsequent experiments and blinding at the gene mutation level. All patients signed informed consent. The study was approved by the Institutional Ethics Committee of the Affiliated Cancer Hospital of Zhengzhou University (No. 2017407). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). All the experiments were completed in the Department of Molecular Pathology of the Affiliated Cancer Hospital of Zhengzhou University.
Design principle and device diagram of the BIOCO collector
The reasons for TAT >5 days mainly include insufficient specimens in the initial stage of the project. To solve this problem, a new type of biopsy specimen collector, BIOCOTM, was designed.
BIOCO is made of polyvinyl chloride (PVC) material using a one-time pressing and it has applied for a patent for utility model in Chins (patent application number is 201820902335.6). Figure 1 shows a schematic diagram of BIOCO from different angles. The BIOCO tool consists of 2 round magnets on the collector that can be adsorbed on the microtome blade holder, thus making it move arbitrarily. Before slicing, the prepared FFPE specimen is fixed on the microtome. The BIOCO collector is then adsorbed on the knife holder, and the leading edge is aligned with the knife. When slicing, the cut tissue will directly enter the collector, without contaminating the knife holder. After the specimen enters the collector, the upper cover with a buckle is fastened to the collector, and the cover is numbered with a marker. The specimen can be processed in the laboratory, after which samples will be stored or shipped. When the laboratory specimen processing starts, the lower piston of the collector is pulled out and tightly connected to the 1.5 mL centrifuge tube, and the upper cover is flicked to make the wax roll enter the centrifuge tube. If there is a residual specimen on the wall, the tube connecting the collector is vertically placed on the test tube rack, dewaxing liquid is directly added into the collector, and the specimen is dissolved into the centrifuge tube for nucleic acid extraction.

Targeted NGS
Human EGFR, KRAS, BRAF, PIK3CA, ALK, and ROS1 Gene Mutation Detection Kits (semiconductor sequencing; Novogene Co., Ltd., Beijing, China) were used to detect 6 lung cancer-related genes. The NGS platform used in the hospital was the DA8600 [National Medical Products Administration (NMPA) approved].
Routine diagnosis and detection pathways
The current protocol in Henan Cancer Hospital relies on 2 primary specimen sources: (I) in-hospital analyses and delivery of the specimen; (II) analyses and delivery of the specimen from Henan Cancer Hospital branches. The puncture specimens from the hospital undergo paraffin embedding, sectioning, hematoxylin-eosin (H&E) staining, IHC ± FISH, and NGS detection in sequence, which takes approximately 7–16 days for the entire molecular pathology detection of lung cancer (Figure 2A). H&E staining, morphological diagnosis, and release of the test report require approximately 1–3 days. The molecular pathology testing for lung cancer can be started only when the H&E report is sent back to the Department of Molecular Pathology. Consequently, 5–8 natural days are required for NGS testing, and the results are then sent back by mail to the clinic. Overall, it takes 8–14 natural days for routine pathway, from receiving samples to sending routine and molecular pathological diagnosis reports of lung cancer (Figure 2B). Through routine detection pathways, a detection cycle for some patients in the hospital lasts about 15 days, while some get discharged, which prevents them from obtaining the treatment plan in a timely manner, or even makes them miss the best treatment time.
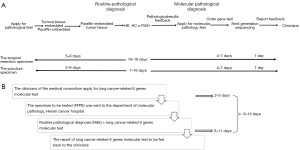
New clinical diagnosis and treatment pathway
TAT refers to the time from when samples are received by the pathology laboratory to when the test results are sent back to the clinicians (4,5). We summed up a new and more efficient diagnosis and treatment pathway with a shorter TAT that is suitable for public hospitals. The protocol (Figure 3) is presented as follows:
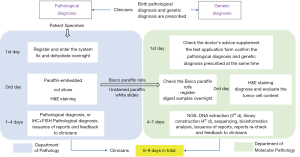
Day 1: according to the relevant examinations and preliminary diagnosis of patients (medical history, routine blood testing, blood coagulation test, electrocardiogram, tumor marker testing, and contrast chest computed tomography data), clinicians propose a pathological and genetic diagnosis (7). The patient’s puncture specimen is sent to the pathology laboratory for registration and entry into the system. The molecular pathology department checks the doctor’s recommendation, communicates with the patient, signs the supplementary application form for the testing of 6 lung cancer-related genes (Novogene Co., Ltd., Beijing, China), and confirms the pathological diagnosis and genetic diagnosis prescribed at the same time.
Day 2: puncture specimens in the pathology department are fixed, dehydrated, embedded in paraffin, cut into paraffin sections, and stained with H&E. At the same time, part of the sample is sent to the molecular pathology department by the medical coordinator. At the Department of Pathology, the diagnosis of IHC and FISH is judged according to the preliminary pathological diagnosis results. If further diagnosis of IHC or FISH is needed, the pathological results are uploaded into the electronic system on the 6th day, and the clinicians are informed. BIOCO paraffin rolls and unstained white paraffin slides are sent to the Department of Molecular Pathology, checked, and registered in the system. BIOCO samples and white films, which are sent to the Department of Molecular Pathology, are also subjected to H&E staining and morphological diagnosis, and the results are checked. When the tumor cell content is greater than 20%, the next detection step is performed.
Day 3–7: the fastest time that NGS can deliver results is in 4 days. On the third day, tissue specimens are dewaxed and digested, and DNA and RNA extraction are performed, followed by RNA reverse transcription. Library construction is performed on day 4. After RNA reverse transcription, the cDNA and extracted DNA are amplified by multiple PCR, followed by primer digestion, amplicon ligation, magnetic beads purification, and library quantification. Consequently, emulsion PCR is performed. On day 5, template enrichment, sequencer cleaning, on-line sequencing, and automatic analysis of biological information are conducted. The generation and checking of the 6 lung cancer-related genes tests are performed on day 6. The report is then fed back to the clinic on day 7. If there is insufficient sample size or other circumstances, the NGS detection period will be extended.
For small biopsy specimens in our hospital, the clinicians prescribe the 6 lung cancer-related genes tests that can be performed within 1 day and the NGS test that takes 4–7 days. The test report is reviewed and fed back to the clinic in 1 day. The entire process lasts 6–9 days.
DNA extraction and quantification
Tumor tissue DNA was extracted from FFPE specimens. DNA extraction was performed using QIAamp DNA FFPE Tissue Kit (Qiagen, Germany, #56404) according to the manufacturer’s instructions. All DNA samples were stored at −20 °C.
Samples were quantified using the NanoDrop 2000 (Thermo Fisher Scientific, USA) and Qubit dsDNA high sensitivity assay kit (Invitrogen by Thermo Fisher Scientifc, Carlsbad, CA, USA, #Q32854) to determine DNA concentrations for the reaction setup. DNA purity was measured by OD260/280 ratio. The appropriate OD260/280 ratio for assessing the purity of DNA is between 1.6 and 2.0.
EGFR gene mutation detection
Quantitative mutation detection was carried out using ACCBio’s human EGFR gene mutation quantitative detection kit (Beijing ACCB Biotech Ltd. #180626), according to the manufacturer’s protocol. Quantitative RT-PCR was performed on an Mx3000P PCR instrument (Agilent, Santa Clara, CA, USA) with the following settings: 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min.
Statistical analysis
T-test was applied for comparison of DNA extraction efficiency and EGFR testing accuracy between new protocol based on BIOCO and the conventional process based on the Routine Collection (ROUCO). All statistical analyses were conducted using the SPSS statistical software, version 19.0 (SPSS Inc., Chicago, IL, USA). P<0.05 was considered statistically significant.
Results
DNA extraction efficiency between BIOCO and ROUCO
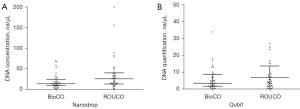
When using the Nanodrop2000, the median DNA concentration of the samples collected by BIOCO was 13.8 ng/µL (1.4–68.5 ng/µL), while that of the ROUCO method was 25.7 ng/µL (2.5–200 ng/µL). When using the Qubit test, the median DNA concentration of the samples collected by BIOCO was 3.5 ng/µL (0.3–34.0 ng/µL), while that of the ROUCO method was 4.3 ng/µL (0.2–27.0 ng/µL) (Figure 4). There was no significant difference (P>0.05) in DNA extraction efficiency between the 2 groups under the same elution volume, which suggested that BIOCO could meet the needs of subsequent testing.
The effect of pollution prevention on BIOCO
Ten paraffin blocks with a known high abundance of EGFR exon 21 L858R mutation (A1-10) and 10 paraffin blocks (B1-10) of EGFR wild type specimens were selected. Alternate sectioning of mutant and wild type paraffin blocks (A1-B1-A2-B2-A3-B3.............A10-B10) was performed. The ROUCO method was used for the A1-10 blocks, and the BIOCO method was used for the B1-10 blocks. Disposable blades and tweezers were replaced before slicing of each paraffin block, and the tool holder was not processed. The cut paraffin was collected into a 1.5 mL centrifuge tube, and DNA was extracted from all 20 specimens according to the standard nucleic acid extraction process, followed by qualitative detection of the EGFR gene. As shown in Table 1, all samples obtained the expected results, and no cross-contamination occurred.
Table 1
| Paraffin block no. | EGFR type | Paraffin block no. | EGFR type |
|---|---|---|---|
| A1 | L858R | B1 | Wild type |
| A2 | L858R | B2 | Wild type |
| A3 | L858R | B3 | Wild type |
| A4 | L858R | B4 | Wild type |
| A5 | L858R | B5 | Wild type |
| A6 | L858R | B6 | Wild type |
| A7 | L858R | B7 | Wild type |
| A8 | L858R | B8 | Wild type |
| A9 | L858R | B9 | Wild type |
| A10 | L858R | B10 | Wild type |
BIOCO, biology collector.
Extraction of DNA by BIOCO and ROUCO for EGFR mutation detection
A total of 39 specimens were collected by the BIOCO and ROUCO methods and were tested for EGFR by the amplification refractory mutation system (ARMS-PCR). The positive rate was 51.3% in both groups (Table 2). The results were completely consistent in terms of the mutant subtypes of EGFR between the 2 groups. We defined 0–10% as low abundance, 10–20% as medium abundance, and >20% as high abundance. The consistencies of mutation with high abundance, medium abundance, and low abundance in the 2 groups were 85.7%, 100%, and 83.3%, respectively. Although the abundance results of the 2 groups were inconsistent, the difference between the 2 groups was not significant (P>0.05).
Table 2
| EGFR mutation detection result | BIOCO (N=39) | ROUCO (N=39) | Consistency (%) |
|---|---|---|---|
| EGFR-WT (case) | |||
| No abundance | 19 (48.7%) | 19 (48.7%) | 100 |
| EGFR-MT (case) | |||
| High abundance | 7 | 6 | 85.7 |
| Medium abundance | 8 | 8 | 100 |
| Low abundance | 5 | 6 | 83.3 |
| Total | 20 (51.3%) | 20 (51.3%) | 100 |
BIOCO, biology collector; ROUCO, Routine Collection.
Real-world TAT data obtained using the new diagnostic and therapeutic pathway
The proportion of in-hospital reports with a TAT <5 days was 72.40%, and the proportion of reports with a TAT of 5–7 days was 14.84%. The proportion of test reports with a TAT within 7–10 days was 12.76%. Therefore, all reports could be issued within 10 days, which provides an additional 4–6 days for patient treatment compared with the routine pathway (Table 3).
Table 3
| Items | Number of specimens | TAT within 5 days/proportion | TAT within 7 days/proportion | TAT within 10 days/proportion |
|---|---|---|---|---|
| Data | 384 | 278/72.40% | 335/87.24% | 384/100% |
TAT, turnaround time.
Discussion
In our hospital, most of the specimens for genetic testing are small biopsy specimens. In addition, the samples used for genetic testing are usually the remaining material from routine pathological diagnosis, so the sample size or the amount of extracted DNA may be insufficient. Our study showed that for about 12% of cases it was not possible to obtain enough specimens, thus many patients needed to undergo a second biopsy. The lack of samples may increase the probability of secondary biopsy, the risk of puncture complications, and places an additional financial burden, as well as reduce the availability of clinical examinations, thus missing the best time for targeted therapy (8).
We found that the main factors affecting TAT included: specimen type, transport, clinical diagnosis and treatment pathway, molecular pathology detection duration, and report release route. In response to the above problems, Henan Cancer Hospital proposed a new diagnosis and treatment protocol. For the cycle problem, we used the Life sequencing platform, which has the fastest detection cycle compared to other sequencers. At the same time, we optimized the diagnosis and treatment pathway to shorten unnecessary waiting times in the process. For the problem of insufficient sample size, we designed a small sample collector, BIOCO, which is used to collect waste samples when constructing wax blocks, and as a supplement when the sample is insufficient. BIOCO has the following advantages: firstly, it could collect the wax rolls that fall when slicing in order to obtain enough sample; secondly, it might reduce the cross-contamination between samples; thirdly, it would simplify the preservation process of the wax rolls samples; fourth, the quality of DNA extraction in BIOCO process is not inferior to the conventional process; at last, effectively improve the reporting cycle of gene testing. The new clinical diagnosis and treatment approach based on Life sequencer and BIOCO collector for public hospitals can provide patients with more timely test reports and accurate treatment. The new clinical diagnosis and treatment approach based on the Life sequencer and BIOCO collector for public hospitals can provide patients with more timely test reports and accurate treatment.
DNA contamination between specimens is also one of the key factors affecting the accuracy of detection (9). According to the requirements of the IASLC guidelines, the slicing knife should be replaced when slicing each specimen and the knife holder and utensils should be wiped with alcohol to prevent cross-contamination. This further increases the burden of experimenters in practical work. The design of BIOCO solves the problem of sample collection and preservation during the trimming of paraffin blocks, simplifies the follow-up workflow of molecular pathology detection, and reduces cross-contamination.
Compared to the ROUCO collection method, our new BIOCO approach showed similar DNA extraction efficiency. The qualitative results of 39 cases found that 1 high-abundance patient obtained a medium-abundance result, and 1 medium-abundance patient obtained a low-abundance result, which may be due to the differences in the total amount of DNA collected or the heterogeneity of the tumor. Overall, about 95% (37/39) of the samples obtained consistent abundance, and 100% (39/39) of the samples obtained consistent mutation detection results, which suggested that the results obtained by the BIOCO collected samples were reliable. From the perspective of the sample amount, the BIOCO collector could improve the success rate of molecular detection.
Of course, there are some aspects of BIOCO that need to be improved. Firstly, as the current BIOCO device is composed of two independent components, the upper cover with a buckle and the collector with two round magnets, this may be inconvenient to operate. In the process of further improvement, it would be improved into an integrated device to make it easier to operate. Secondly, at present the BIOCO device could not control the number of wax rolls we need. We hoped that the improved device could more accurately release the specified number of wax rolls according to the needs of various gene testing.
However, this study has a few limitations. In order to shorten the period of molecular pathology detection, molecular pathology H&E staining was simultaneously performed with overnight digestion of the samples. If H&E staining showed no tumor cells or the content of tumor cells was less than 20%, it did not meet the criteria of pathological quality control, and tumor tissue enrichment was required.
Conclusions
The new diagnosis and treatment pathway accelerates the reporting period of molecular pathology detection, shortens the TAT to 4–6 days, and provides the opportunity for rapid and precise treatment of patients. At the same time, the rapid and accurate molecular pathology detection report accelerated the turnover of the hospital beds, improved the efficiency of the use of hospital beds, and reduced the medical expenses. The DA8600 platform and BIOCO collector were shown to be effective tools for clinicopathological diagnosis, which can be used by pathology and related departments.
Acknowledgments
We thank MedSci for proofreading the manuscript.
Funding: This study was supported by the National Natural Science Foundation of China (81802444) and Science and Technology Department of Henan Province (212102310128).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6230/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6230/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6230/coif). XL, LM, and YY are from Novogene Co., Ltd., Beijing, China. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). All patients signed informed consent. The study was approved by the Institutional Ethics Committee of the Affiliated Cancer Hospital of Zhengzhou University (No. 2017407).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cao L, Long L, Li M, et al. The utilization of next-generation sequencing to detect somatic mutations and predict clinical prognosis of Chinese non-small cell lung cancer patients. Onco Targets Ther 2018;11:2637-46. [Crossref] [PubMed]
- Lung Cancer Group, Respiratory Society, Chinese Medical Association. Expert consensus on molecular targeted therapy for advanced NSCLC (2013). Chinese Journal of Tuberculosis and Respiratory Diseases 2014;37:177-82.
- NCCN. The NCCN Non-small Cell Lung Cancer clinical practice guidelines in oncology (version 1.2021). Available online: http://www.nccn.org
- Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med 2013;137:828-60. Erratum in: Arch Pathol Lab Med 2013;137:1174. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol 2018;13:323-58. [Crossref] [PubMed]
- DiStasio M, Chen Y, Rangachari D, et al. Molecular Testing Turnaround Time for Non-Small Cell Lung Cancer in Routine Clinical Practice Confirms Feasibility of CAP/IASLC/AMP Guideline Recommendations: A Single-center Analysis. Clin Lung Cancer 2017;18:e349-56. [Crossref] [PubMed]
- Jiang T, Ren S, Li X, et al. The changing diagnostic pathway for lung cancer patients in Shanghai, China. Eur J Cancer 2017;84:168-72. [Crossref] [PubMed]
- Atanesyan L, Steenkamer MJ, Horstman A, et al. Optimal Fixation Conditions and DNA Extraction Methods for MLPA Analysis on FFPE Tissue-Derived DNA. Am J Clin Pathol 2017;147:60-8. [Crossref] [PubMed]
- Shen LS, Xie GH. Molecular diagnosis in China: present status and challenges ahead. Chinese Journal of Laboratory Medicine 2016;39:473-6.
(English Language Editor: C. Betlzar)


