
This article has an erratum available at: http://dx.doi.org/10.21037/atm-2022-18 the article has been update on 2022-06-10 at here.
Modification of STIM2 by m6A RNA methylation inhibits metastasis of cholangiocarcinoma
Introduction
Cholangiocarcinoma (CCA) is the second most aggressive form of primary liver cancer, with an increasing incidence worldwide (1,2). CCA has gradual signs, a high degree of malignancy, and a poor prognosis, as several parts of its clinical features are identical to those of hepatocellular carcinoma (HCCA) (3). Radiation therapy, chemotherapy, intervention, surgery, immune targeting, CCA ablation treatment, and other multidisciplinary approaches are all options for treating CCA (4-7). CCA is generally unresponsive to radiotherapy and chemotherapy, and consequently has inferior clinical outcomes.
CCA is a highly fatal malignant tumor originating from the bile duct epithelium, which is characterized by late clinical manifestations and lack of effective treatment. Chronic inflammation, including primary sclerosing cholangitis, Fasciola hepatica infection and hepatolithiasis, is listed as a risk factor, but the cause is unclear for most cases of CCA. The latest progress in molecular pathogenesis emphasizes the apparent legacy. The importance of genetic changes, including promoter hypermethylation and histone deacetylation, and genetic changes in cholangiocarcinoma (8). N6-methyladenosine (m6A) is a common RNA (ribonucleic acid) alteration that predictably affects carcinogenesis and tumor growth (9). The alteration of m6A RNA is a reversible process that is coordinated by methyltransferase (m6A “writers” and “readers”) and demethylase (m6A “erasers”) proteins (10-12). Recent research indicates that m6A modification may play a critical role in various malignant cancers, including CCA (13-16). However, the particular chemical mechanism by which m6A induces CCA development remains largely unknown.
Orai1 are essential primary proteins that are clustered and activated by STIM1 (Stromal Interaction Molecule 1) and STIM2 (Stromal Interaction Molecule 2). STIM1 is the direct activator of Orai1, and loss of the protein eliminates Orai1 activation, thereby regulating downstream Ca2+ (calcium)-dependent cell functions (17). STIM1 clustering of Orai1 in endoplasmic reticulum (ER)-plasma membrane (PM) junctions reinforces the coupling of Orai1-mediated Ca2+ entry with nuclear factor of activated T cells 1 (NFAT1) initiation (18). In contrast, the activation of diffusely localized channels in the membrane by the cytosolic STIM1 C terminus or SOAR (the STIM1 Orai activating region) fragments has been demonstrated in some research to cause nuclear translocation of NFAT1 and gene expression (17-20). STIM2 is also considered to be a relevant participant in pathological conditions associated with aging, Alzheimer’s and Huntington’s diseases, autoimmune diseases and cancer (21). STIM2 is considered to play a role in tumor development control; however, research on this is limited. However, the role of STIM2 in CCA regulation remains unclear. The purpose of this study was to investigate the clinical significance and biological roles of STIM2 in CCA.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6485/rc).
Methods
Samples and cell lines from human origin
The Eastern Hepatobiliary Surgery Hospital (EHBH) used CCA tissues and neighboring normal tissues from 10 CCA patients undergoing curative surgical resections. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Review Committee of Eastern Hepatobiliary Surgery Hospital and all patients provided written informed consent. Human CCA cell lines HuCCAT1 (ATCC, Manassas, USA)were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium with 100 g/mL streptomycin, 100 U/mL penicillin, and 10% fetal calf serum (GE Healthcare, Life Sciences, USA). The third-party biology services used short tandem repeat (STR) analysis to characterize all cell lines (Feiouer Biology Co., Ltd., Chengdu, China).
RT and qPCR (reverse transcription-reaction and quantitative polymerase chain reaction)
The PrimeScript RT reagent kit (Takara Bio Inc., China) produced the first-strand cDNA (complementary DNA). To synthesize microRNA (miRNA), the first-strand cDNA Poly-A Tailing Kit (Sangon Bio Inc., Shanghai, China) was used to reverse-transcribe miRNAs. In a quantitative RT-PCR, SYBR Premix Ex TaqII (Takara Bio Inc., Dalian, China) was utilized (qRT-PCR). The endogenous controls for Messenger RNA (mRNA) and miRNAs were β-actin and U6, respectively. The relative fold-change of target expression was calculated using the comparative 2-ΔΔCt technique. Sangon designed and manufactured all of the primers (Shanghai, China).
Immunohistochemical (IHC) staining
IHC was performed according to the previously described method (22). The evaluation fields were set at 200× magnification power for H-score calculations (23). In malignant cells, staining concentration was sorted as 0, 1, 2, or 3, referring to the existence of negative, weak, intermediary, or solid brown staining, respectively. All of the cells were counted and stained at different intensities to obtain an overall cell count. To calculate the average percentage of positive cells, we used the formula shown below.
H-score = (% of cells stained at intensity level 1×1) + (% of cells stained at intensity level 2×2) + (% of cells stained at intensity level 3×3). H-scores of 0–3 were obtained, with 0 indicating that all cells stained negatively and 3 indicating that all cells stained positively.
Establishment of stable cell lines overexpressing STIM2 and Keratin 8 (KRT8)
Hanbio provided the lentivirus vectors expressing STIM2 or KRT8 (LV-STIM2 or LV-KRT8) and its control vector (LV-NC) (Shanghai, China). CCA cells were infected with the lentivirus vector and then treated with puromycin (at a 2 µg/mL concentration) for 1 week.
In vivo experimentation through metastasis assays
A protocol was prepared before the study without registration. Animal experiments were approved by the Second Military Medical University’s Institutional Animal Care and Use Committee (Shanghai, China), in compliance with the Second Military Medical University guidelines for the care and use of animals. Four-week-old male BALB/c mice were purchased from the Second Military Medical University Animal Center and maintained in pathogen-free conditions with a regular pellet diet and water. Metastatic lung and liver models were used as previously described (23).
Methylated RNA immunoprecipitation (MeRIP) PCR
MeRIP-PCR analysis was performed according to a previously published technique (24).
Statistical analysis
The mean± standard error of the mean (SEM) were used to present the data. All data were obtained from three independent experiments. GraphPad Prism v.8 (San Diego, USA) and SPSS v.25.0 (Chicago, USA) software were used for all statistical analyses. Survival curves were calculated using the Kaplan-Meier method, and the differences were determined using the log-rank test. Univariate and multivariate Cox proportional hazards regression models were used to examine the independent components. P values less than *0.05, **0.01, and ***0.001 were deemed statistically significant.
Results
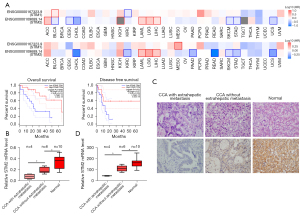
STIM2 deficiency in CCA tissues is related to a poor prognosis and extrahepatic metastases
According to the GEPIA(Gene Expression Profiling Interactive Analysis) 2 database, patients with CCA who had lower levels of STIM2 had a worse overall prognosis and were more likely to die from the condition (Figure 1A). STIM2 mRNA expression was significantly lower in 10 matched CCA tissues compared to adjacent normal tissues by quantitative PCR (qPCR). Notably, the CCA that metastasized extra hepatically had the lowest STIM2 mRNA expression (Figure 1B). Meanwhile, IHC revealed that STIM2 protein levels were commensurate with the trend in mRNA levels (Figure 1C,1D). Collectively, these findings suggested that STIM2 may be involved in the evolution of CCA and may serve as a possible prognostic indicator for patients with CCA.
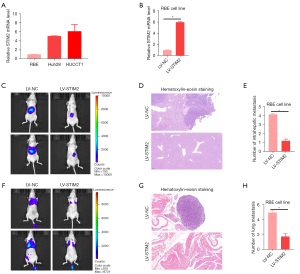
Inhibition of tumor metastasis by STIM2 in vivo
The mRNA level of STIM2 was then studied in three CCA cell lines, with RBE exhibiting relatively lower STIM2 expression, while HUCCAT1 and Huh28 exhibited markedly higher STIM2 expression (Figure 2A). To investigate the role of STIM2 in preventing CCA cell metastasis, RBE cells were infected with LV-STIM2 or LV-NC (Figure 2B), and the effects of STIM2 on tumor metastasis were examined in vivo. To create the liver orthotopic-implanted models, cells were infected with LV-STIM2 or LV-NC and transplanted into the livers of nude mice. Six weeks after transplantation, the fluorescence signal intensities of metastatic liver nodules in the LV-STIM2 group were significantly lower than those in the LV-NC group (Figure 2C). There were significantly fewer metastatic foci found in liver tissue slices from patients who had been treated with LV-STIM2 (Figure 2D,2E), indicating that STIM2 inhibited the ability of CCA cells to spread intra-hepatically. Using nude mice as a model for lung metastasis, we injected cells labeled with firefly luciferase into their tail veins. Compared to the LV-NC group, the LV-STIM2 group had significantly lower bioluminescence signal intensities in mice, substantially lower fluorescence signal intensities of metastatic lung nodules (Figure 2F), and fewer metastatic foci in lung tissue sections (Figure 2G,2H), indicating that STIM2 can inhibit hepatoma cells’ metastatic lung potential.
STIM2 inhibits tumor metastasis by decreasing KRT8 levels in vivo
Next, we explored the targeted gene of STIM2 inhibition of tumor metastasis in vivo. The UALCAN database (https://ualcan.path.uab.edu/) showed that the top five genes (KRT18, KRT8, MPST, MRPS17, and C20orf24) were negatively correlated with STIM2 in CCA (Figure 3A). We observed that LV-STIM2 lowered KRT8 mRNA expression in CCA cells, indicating that STIM2 may regulate KRT8 expression in CCA cells (Figure 3B). As a result, we are curious as to whether STIM2 inhibits tumor metastasis via decreased KRT8 levels. SK-Hep1 cells transfected with LV-STIM2 or LV-NC were infected in vitro with LV-KRT8 or LV-N, respectively (Figure 3C). LV-STIM2 decreased the capacity of CCA to metastasize in orthotopic implanted and lung metastasis models. Simultaneously, LV-KRT8 restored the inhibitory effect of STIM2 on CCA cell metastasis in vivo (Figure 3D-3I), demonstrating that STIM2 suppresses tumor metastasis in vivo by lowering KRT8 levels.

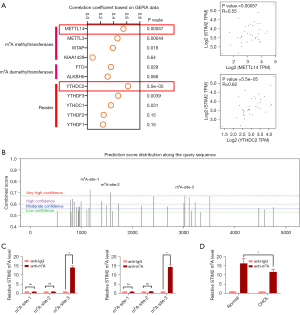
Abnormal STIM2 levels in CCA are responsible for m6A methylation modification
Through GEPIA 2 analysis, we observed the relationship between STIM2 and the expression of methylase METTL3, METTL14, WTAP, and KIAA1429, as well as demethylase ALKBH5 and FTO. In the CCA, we detected that STIM2 was related toMETTL14 and YTHDC2 (Figure 4A). Meanwhile, Figure 4B shows that STIM2 mRNA had three very confident m6A sites, indicating that m6A modification may be related to aberrant STIM2 levels in CCA, as demonstrated by the SRAMP (sequence-based RNA adenosine methylation site predictor) database (https://www.cuilab.cn/sramp). Further research using a RIP (RNA immunoprecipitation) assay confirmed the presence of the m6A-3-site in STIM2 mRNA (Figure 4C). In the clinic, MeRIP-qPCR results showed that in 10 paired CCA patients, the m6A CPEB1 level was lower in the tumors than the surrounding normal tissues (Figure 4D). According to the above findings, aberrant STIM2 levels in CCA are caused by m6A alteration.
Discussion
CCA, which develops from epithelial cells confronting the biliary tree lumen, is the second most common primary hepatic tumor worldwide after HCCA (25). As a result of the lack of early recognized clinical symptoms and the unavailability of precise tumor biomarkers, CCA is typically detected at an advanced, incurable stage (26). Despite recent advances in the development of molecularly targeted medicines, the outlook for this lethal malignancy remains bleak. As a result, there is a high priority in unraveling the molecular mechanisms and pathways underlying this disease to improve the clinical outcomes of CCA patients. STIM2 is the primary protein involved in Orai1 clustering and activation (27).
There is little known regarding the involvement of STIM2 in the advancement of CCA. Based on the GEPIA 2 database, this study discovered that low STIM2 mRNA levels were related to poor survival in CCA patients. Quantitative PCR and IHC studies revealed a decrease in the protein expression in CCA tissues was linked to extrahepatic metastasis. Increased STIM2 levels inhibited CCA cell metastasis in vivo, confirming the inhibitory effect of STIM2 on the tumorigenesis and metastasis of CCA.
Our mechanistic investigation revealed that the STIM2-KRT8 axis is critical in CCA metastasis. KRT8 (keratin 8) is an epithelial marker and one of the most important keratin proteins (28). KRT8 and its filament partner, KRT18, have been found to control the cellular response to stress stimuli and contribute to cell resistance to apoptosis, in addition to maintaining cell mechanical integrity (29). Recent investigations showed that KRT8 levels were higher in the aqueous humor of patients with age-related macular degeneration (AMD) (30). In vitro studies have also indicated that high KRT8 levels could protect RPE cells from deterioration under oxidative stress (31). KRT8 has been demonstrated to be important in a variety of malignant tumors (32-34). However, the molecular mechanism of KRT8 in the pathogenesis of CCA remains unknown. In this study, the GEPIA database and qRT-PCR were used to determine the changed expression of the KRT8 mRNA levels controlled by STIM2. According to functional studies, STIM2 suppresses tumor metastasis in vivo by reducing KRT8 levels. In future research, we will explore the unique regulatory mechanism of m6A-induced STIM2 abnormalities in CCA.
A recent study revealed that m6A mRNA modification is critical for RNA fate, including splicing, localization, translation, transport, and mRNA stability (35). A multicomponent complex composed of METTL3, METTL14, and WTAP mediates adenosine N6 methylation in mammals (36). To date, FTO and ALKBH5 have been shown to influence m6A demethylation (37). m6A has been shown to interact with the targeted mRNA or miRNA and contribute to the progression of various types of cancer (9). In recent years, emerging studies have suggested that m6A-related genes have crucial roles in the initiation and progression of cancers, including acute myeloid leukaemia, renal cell carcinoma, and hepatocellular carcinoma etc. (24). Several m6A regulator abnormal expressed is closely related to the poor prognosis of tumor patients including CCA (38). Recently study identified PD-L1 mRNA as a target of ALKBH5 and reveals a role for ALKBH5 in regulating the tumor immune microenvironment and immunotherapy efficacy in CCA (39). However, research on the m6A alteration of STIM2 in CCA is lacking. In the present study, the SRAMP database and MeRIP-seq (sequencing) were used to determine the altered expression of STIM2 mRNA transcripts controlled by m6A alteration. In this study, we found that the m6A/IGF2BP2 axis functioned in enhancing CCA metastasis, suggesting that it might be a new target for CCA diagnosis and treatment. Finally, as previously stated, this was the first time that STIM2 was identified as a direct target of m6A-mediated transformation. The particular regulatory mechanism through which m6A induces STIM2 abnormality in CCA remains an open question for further research.
Conclusions
Finally, our findings have provided new insight into the critical involvement of the m6A-STIM2 axis in the progression of CCA metastasis. The current results highlighted the importance of the molecular process of m6A epi-transcriptomic alteration in cancer research, which was previously undiscovered.
Acknowledgments
Funding: This study was supported by grants from the Science Fund for Creative Research Groups, NSFC, China (No. 81900550); and the Shanghai Tongren Hospital (No. 2019shtrxx09).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6485/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6485/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6485/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Review Committee of Eastern Hepatobiliary Surgery Hospital and all patients provided written informed consent. Animal experiments were approved by the Second Military Medical University’s Institutional Animal Care and Use Committee (Shanghai, China), in compliance with the Second Military Medical University guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Rizvi S, Khan SA, Hallemeier CL, et al. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat Rev Clin Oncol 2018;15:95-111. [Crossref] [PubMed]
- Sirica AE, Gores GJ, Groopman JD, et al. Intrahepatic Cholangiocarcinoma: Continuing Challenges and Translational Advances. Hepatology 2019;69:1803-15. [Crossref] [PubMed]
- Hoy SM. Pemigatinib: First Approval. Drugs 2020;80:923-9. [Crossref] [PubMed]
- Abou-Alfa GK, Macarulla T, Javle MM, et al. Ivosidenib in IDH1-mutant, chemotherapy-refractory cholangiocarcinoma (ClarIDHy): a multicentre, randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol 2020;21:796-807. [Crossref] [PubMed]
- Sittithumcharee G, Suppramote O, Vaeteewoottacharn K, et al. Dependency of Cholangiocarcinoma on Cyclin D-Dependent Kinase Activity. Hepatology 2019;70:1614-30. [Crossref] [PubMed]
- Simile MM, Bagella P, Vidili G, et al. Targeted Therapies in Cholangiocarcinoma: Emerging Evidence from Clinical Trials. Medicina (Kaunas) 2019;55:42. [Crossref] [PubMed]
- Isomoto H. Epigenetic alterations in cholangiocarcinoma-sustained IL-6/STAT3 signaling in cholangio- carcinoma due to SOCS3 epigenetic silencing. Digestion 2009;79:2-8. [Crossref] [PubMed]
- He L, Li H, Wu A, et al. Functions of N6-methyladenosine and its role in cancer. Mol Cancer 2019;18:176. [Crossref] [PubMed]
- Wang T, Kong S, Tao M, et al. The potential role of RNA N6-methyladenosine in Cancer progression. Mol Cancer 2020;19:88. [Crossref] [PubMed]
- Chen M, Wong CM. The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol Cancer 2020;19:44. [Crossref] [PubMed]
- Zhou Z, Lv J, Yu H, et al. Mechanism of RNA modification N6-methyladenosine in human cancer. Mol Cancer 2020;19:104. [Crossref] [PubMed]
- Chen XY, Zhang J, Zhu JS. The role of m6A RNA methylation in human cancer. Mol Cancer 2019;18:103. [Crossref] [PubMed]
- Bo C, Li N, He L, et al. Long non-coding RNA ILF3-AS1 facilitates hepatocellular carcinoma progression by stabilizing ILF3 mRNA in an m6A-dependent manner. Hum Cell 2021;34:1843-54. [Crossref] [PubMed]
- Xue J, Xiao P, Yu X, et al. A positive feedback loop between AlkB homolog 5 and miR-193a-3p promotes growth and metastasis in esophageal squamous cell carcinoma. Hum Cell 2021;34:502-14. [Crossref] [PubMed]
- Chen YT, Xiang D, Zhao XY, et al. Upregulation of lncRNA NIFK-AS1 in hepatocellular carcinoma by m6A methylation promotes disease progression and sorafenib resistance. Hum Cell 2021;34:1800-11. [Crossref] [PubMed]
- Huang GN, Zeng W, Kim JY, et al. STIM1 carboxyl-terminus activates native SOC, I(crac) and TRPC1 channels. Nat Cell Biol 2006;8:1003-10. [Crossref] [PubMed]
- Park CY, Hoover PJ, Mullins FM, et al. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell 2009;136:876-90. [Crossref] [PubMed]
- Samanta K, Kar P, Mirams GR, et al. Ca(2+) Channel Re-localization to Plasma-Membrane Microdomains Strengthens Activation of Ca(2+)-Dependent Nuclear Gene Expression. Cell Rep 2015;12:203-16. [Crossref] [PubMed]
- Kawasaki T, Lange I, Feske S. A minimal regulatory domain in the C terminus of STIM1 binds to and activates ORAI1 CRAC channels. Biochem Biophys Res Commun 2009;385:49-54. [Crossref] [PubMed]
- Berna-Erro A, Jardin I, Salido GM, et al. Role of STIM2 in cell function and physiopathology. J Physiol 2017;595:3111-28. [Crossref] [PubMed]
- Zheng H, Bi FR, Yang Y, et al. Downregulation of miR-196-5p Induced by Hypoxia Drives Tumorigenesis and Metastasis in Hepatocellular Carcinoma. Horm Cancer 2019;10:177-89. [Crossref] [PubMed]
- Zheng H, Yang Y, Hong YG, et al. Tropomodulin 3 modulates EGFR-PI3K-AKT signaling to drive hepatocellular carcinoma metastasis. Mol Carcinog 2019;58:1897-907. [Crossref] [PubMed]
- Chen JQ, Tao YP, Hong YG, et al. M6A-mediated up-regulation of LncRNA LIFR-AS1 enhances the progression of pancreatic cancer via miRNA-150-5p/ VEGFA/Akt signaling. Cell Cycle 2021;20:2507-18. [Crossref] [PubMed]
- Huang L, Jiang X, Li Z, et al. Linc00473 potentiates cholangiocarcinoma progression by modulation of DDX5 expression via miR-506 regulation. Cancer Cell Int 2020;20:324. [Crossref] [PubMed]
- Singsuksawat E, Thuwajit C, Charngkaew K, et al. Increased ETV4 expression correlates with estrogen-enhanced proliferation and invasiveness of cholangiocarcinoma cells. Cancer Cell Int 2018;18:25. [Crossref] [PubMed]
- Pan X, Wang G, Wang B. MicroRNA-1182 and let-7a exert synergistic inhibition on invasion, migration and autophagy of cholangiocarcinoma cells through down-regulation of NUAK1. Cancer Cell Int 2021;21:161. [Crossref] [PubMed]
- Miao Q, Xu Y, Yin H, et al. KRT8 phosphorylation regulates the epithelial-mesenchymal transition in retinal pigment epithelial cells through autophagy modulation. J Cell Mol Med 2020;24:3217-28. [Crossref] [PubMed]
- Tao GZ, Looi KS, Toivola DM, et al. Keratins modulate the shape and function of hepatocyte mitochondria: a mechanism for protection from apoptosis. J Cell Sci 2009;122:3851-5. [Crossref] [PubMed]
- Kang GY, Bang JY, Choi AJ, et al. Exosomal proteins in the aqueous humor as novel biomarkers in patients with neovascular age-related macular degeneration. J Proteome Res 2014;13:581-95. [Crossref] [PubMed]
- Baek A, Yoon S, Kim J, et al. Autophagy and KRT8/keratin 8 protect degeneration of retinal pigment epithelium under oxidative stress. Autophagy 2017;13:248-63. [Crossref] [PubMed]
- Fang J, Wang H, Liu Y, et al. High KRT8 expression promotes tumor progression and metastasis of gastric cancer. Cancer Sci 2017;108:178-86. [Crossref] [PubMed]
- Xie L, Dang Y, Guo J, et al. High KRT8 Expression Independently Predicts Poor Prognosis for Lung Adenocarcinoma Patients. Genes (Basel) 2019;10:36. [Crossref] [PubMed]
- Scott MKD, Limaye M, Schaffert S, et al. A multi-scale integrated analysis identifies KRT8 as a pan-cancer early biomarker. Pac Symp Biocomput 2021;26:297-308. [PubMed]
- Zhu W, Wang JZ, Wei JF, et al. Role of m6A methyltransferase component VIRMA in multiple human cancers Cancer Cell Int 2021;21:172. (Review). [Crossref] [PubMed]
- Wang J, Chen L, Qiang P. The role of IGF2BP2, an m6A reader gene, in human metabolic diseases and cancers. Cancer Cell Int 2021;21:99. [Crossref] [PubMed]
- Wang JY, Lu AQ. The biological function of m6A reader YTHDF2 and its role in human disease. Cancer Cell Int 2021;21:109. [Crossref] [PubMed]
- Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother 2019;112:108613. [Crossref] [PubMed]
- Qiu X, Yang S, Wang S, et al. M6A Demethylase ALKBH5 Regulates PD-L1 Expression and Tumor Immunoenvironment in Intrahepatic Cholangiocarcinoma. Cancer Res 2021;81:4778-93. [Crossref] [PubMed]


