
CLCF1 is up-regulated in renal ischemia reperfusion injury and may associate with FOXO3
Introduction
Renal ischemia-reperfusion injury (IRI), an inevitable event during kidney transplantation, is recognized as one of the key factors causing delayed graft function (DGF) (1). DGF is a common complication after kidney transplantation and is associated with decreased short-term and long-term graft survival rates. Ischemic injury occurs following the clamping of blood flow from the donor, which causes necrosis and tissue damage. The reperfusion of blood from the recipient augments the graft injury (2,3). In general, oxidative and inflammatory reactions play a key role in renal IRI.
IRI contributes to DGF along with multiple factors, including the specific characteristics of the donor, recipient, and operation (1). However, the severity of IRI does not necessarily correlate with the occurrence of DGF (4). This fact suggests that IRI and other risk factors contribute to DGF to varying degrees, either independently or jointly, which makes the etiological diagnosis of DGF difficult. Confusion regarding the primary cause of DGF is not conducive to prognostic judgment and accurate treatment. While IRI is recognized as one of the most important risk factors of DGF (1), the mechanism that links IRI to DGF remains unclear. Until recently, no clear “bridge” molecule linking IRI to DGF has been found, which has hindered the search for the mechanism of progression from IRI to DGF. Prior to this study, we screened for differentially expressed genes in a murine model of renal IRI induced by clamping renal vessels. We found 2,218 differentially expressed genes in the IRI-treated kidney tissues (5,6). When we compared these results with the DGF database (published by Mueller) (7), we found 107 overlapping genes, of which cardiotrophin-like cytokine factor 1 (CLCF1) was one of the most upregulated.
CLCF1 is a secretory cytokine of the interleukin-6 (IL-6) family. CLCF1 binds to the cytokine receptor-like factor 1 (CRLF1) or soluble ciliary neurotrophic factor receptor (sCNTFR) (8). CLCF1/CRLF1 or CLCF1/sCNTFR forms a signaling complex on the cell membrane and activates the signal transducer and activator of transcription-3 (STAT3) pathway (9). CLCF1 has been reported to play important roles in many diseases, such as cold-induced sweating syndrome (CISS) (10), neurodegenerative diseases (11), and more recently, idiopathic focal segmental glomerulosclerosis (FSGS) (12,13).
However, the regulation of CLCF1 expression, especially in renal IRI, is not clear. The forkhead box O3 (FOXO3) gene is a known transcription factor associated with IL-6 (13). In FOXO3-deficient mice, the expression of IL-6 is increased, and FOXO3 inhibits IL-6 production in the dendritic cells (DCs) of wild type mice (13). Both IL-6 and CLCF1 belong to the IL-6 cytokine family. Therefore, we speculated that CLCF1 might also be regulated by FOXO3. In the present study, we investigated the expression and regulation of CLCF1 in an IRI mouse model. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-4381/rc).
Methods
Animal study
Male C57BL/6 mice (8–10-week-old, 21–23 g) were used in this study. All mice were kept under specific pathogen-free (SPF) conditions and allowed free water and food access (14 h light/10 h dark cycle, 20–21 ℃ temperature, humidity: 60%). The animal experiments in this study were approved by the Beijing Friendship Hospital Animal Care and Use Committee (No. 18-2022). A protocol was prepared before the study without registration. In this study, all procedures involving animals were performed and monitored in compliance with the guidelines of Beijing Friendship Hospital Animal Use Regulations. The wild type mice (n=40) were obtained from Vital River Laboratories (Beijing, China). Global FOXO3 knockout mice (n=16) were generated from Biocytogen (Beijing, China) as described previously (14). Mice were weighed and randomly distributed into 4 groups: (I) Sham-WT (wild type, sham operation); (II) Injury-WT (wild type, kidney IRI operation); (III) Sham-KO (FOXO3 knockout, sham operation); (IV) Injury-KO (FOXO3 knockout, kidney IRI operation).
The renal IRI operation was performed as previously described (2). Briefly, the mice were anesthetized with pentobarbital sodium (0.05 mg/g body weight) through intraperitoneal injection. The kidneys of both sides were exposed. To induce IRI, the kidney arteries were clamped for 45 min of ischemia and reperfused for 24 h. The mice were sacrificed by carbon dioxide euthanasia, and the samples were immediately collected and stored at −80 ℃. Sham-operated mice were used as controls. Mouse serum creatinine (Cr) is detected using a Creatinine Assay (sarcosine oxidase) kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The procedure was performed according to the manufacturer’s instructions. In brief, 6 µL of serum samples, standard Cr solution, or deionized water were mixed with 180 µL of Enzyme A solution and incubated at 37 ℃ for 5 min. The optical density (OD) at 546 nm (A1) was obtained with a microplate reader. Then, 60 µL of Enzyme B solution was added and incubated at 37 ℃ for 5 min, the OD at 546 nm was read (A2), and the serum Cr was calculated according to Cr (µmol/L) = ((ΔA (sample)-ΔA (blank))/(ΔA (standard)-ΔA (blank)))×C (standard). ΔA=A2- K×A1, K=( (Volume(Enzyme A solution)+ Volume(sample) )/ ( (Volume(Enzyme A solution)+ Volume(sample)+Volume(Enzyme B solution) ), C (standard) = 442µmol/L. The animal experiments were carried out in an SPF environment in the animal laboratory in Beijing Friendship Hospital.
Histological analysis
The fixed tissues were routinely dehydrated, embedded in paraffin, and cut into 4 µm sections. Routine hematoxylin and eosin (H&E) staining was performed for histological analysis. For immunohistochemistry (IHC), the sections were subjected to an antigen retrieval process, followed by incubation with 3% of H2O2 for 20 min to block endogenous peroxidase. The sections were incubated with diluted rabbit anti-CLCF1 antibody (Novus Biologicals, CO, USA) at 4 ℃ overnight. The sections were then washed 3 times with phosphate-buffered saline (PBS) and incubated with horseradish peroxidase (HRP)/Fab polymer-conjugated secondary antibody (ZSGB-Bio, Beijing, China) for 1 h at room temperature. The expression of CLCF1 was visualized with the diaminobenzidine (DAB) assay (ZSGB-Bio) and sections were counterstained with hematoxylin. The sections were viewed using a microscope.
Quantitative real-time PCR assay
Total RNA was quantified and reverse transcribed using a cDNA Synthesis Kit (Takara, Dalian, China). The expression of specific genes was determined using a real-time qPCR assay. The primers used were as follows: mouse CLCF1 forward, AAACCTATGACCTCACCCGC; reverse, TTGAGGCCACGCAAGTAACA. GAPDH was used as an internal control to normalize the expression levels: forward, GGCATTGTGGAAGGGCTC and reverse: GGGGGTAGGAACACGGAAG. The thermal reaction was: 95 ℃ for 30 s, 40 repetitions of 95 ℃ for 5 s and 60 ℃ for 30 s.
Western blot
Kidney tissues were homogenized and quantified. A total of 50 µg protein was loaded onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for electrophoretic separation. The proteins were then blotted onto the nitrocellulose membrane and blocked with 5% non-fat milk at room temperature for 2 h. The membrane was then incubated with rabbit anti-FOXO3 (Abcam, MA, USA) or mouse anti-β-actin (Santa Cruz, CA, USA) antibodies at 4 ℃ overnight. Subsequently, the membrane was washed 4 times using Tris-buffered solution with Tween-20 (TBST), followed by incubation with HRP-conjugated goat anti-rabbit or goat anti-mouse antibodies for 1 h at room temperature. The membrane was then washed 4 times with TBST, and the specific bands were visualized with SuperSignal West Femto Maximum Sensitivity Substrate (Pierce, IL, USA).
Enzyme-linked immunosorbent assay
CLCF1 in the mouse serum samples were measured using double antibody sandwich method mouse enzyme-linked immunosorbent assay (ELISA) kits (Cusabio, Wuhan, China). The procedure was performed according to the manufacturer’s instruction. Briefly, serum samples were loaded onto an antibody pre-coated 96-well plate and incubated at 37 ℃ for 2 h. Subsequently, the supernatants were replaced with a biotin-conjugated antibody and incubated at 37 ℃ for 1 h. The wells were washed 5 times with a washing solution, then incubated with HRP-conjugated avidin at 37 ℃ for 1 h. The plate was then washed 5 times, and the tetramethylbenzidine (TMB) substrate was applied for reaction at 37 ℃ for 1 h. The reaction was stopped by the addition of a stop solution, and the OD at 450 nm was recorded using a microplate reader. The levels of CLCF1 were calculated according to the standard curve.
Statistical analysis
Data are expressed as means ± SEM. All statistical analysis was performed using SPSS 19.0 software (SPSS Inc., Chicago, IL, USA). The differences between the 2 groups were analyzed using a Student’s t-test. Cr data comparisons were performed using analysis of variance (ANOVA) with nonparametric Games-Howell post hoc analysis. A P value of less than 0.05 was considered statistically significant.
Results
Data overlap and bioinformatic analysis
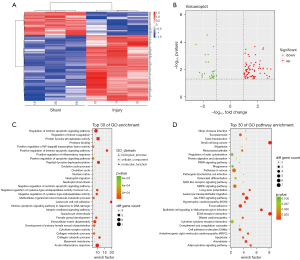
To explore the differentially expressed genes in renal IRI, a mouse bilateral renal artery clamping model was constructed, and the expression profiles (Shanghai Biotechnology Corporation, Shanghai, China) were screened using the Mouse Genome 430 2.0 microarray (Thermo Fisher Scientific) (5,6). Data from the high-throughput screening identified 2218 differentially expressed genes, including 1,103 up-regulated and 1,115 down-regulated genes (P<0.05). The DGF database (7), based on graft biopsies of 11 DGF recipients and 76 immediate graft function (IGF) recipients, was published by Mueller and included a total of 1,109 transcripts that differed between DGF and IGF (unadjusted P value <0.05). By comparing the IRI model database with the DGF database, we identified 107 overlapped and co-directional genes (Figure 1A,1B, Tables S1,S2). We performed gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the overlapped genes (Figure 1C,1D, Table S2) and found that apoptosis, DNA damage, and inflammation were major pathways in the 107 genes. Among the 107 overlapped genes, we found that CLCF1 was up-regulated to 11.1-fold in the IRI mouse model (IRI group vs. sham group, P<0.001) and 1.3-fold in the DGF database (DGF group vs. IGF group, unadjusted P value =0.0134).
CLCF1 is up-regulated by renal IRI
To verify whether CLCF1 expression could be up-regulated by renal IRI, we performed a real-time qPCR assay of the renal tissues of IRI-treated mice. We observed that CLCF1 was indeed up-regulated in the IRI-treated mice compared with the sham-operated control group (Figure 2A, Table S2). IHC staining data indicated that CLCF1 was strongly accumulated around the injured kidney tubules, whereas CLCF1 showed weak expression in sham operated kidney in the kidney tissues of the sham-operated mice (Figure 2B, Table S2).

As CLCF1 is a secretary factor, we detected circulatory CLCF1 in the serum samples. We found that serum CLCF1 was significantly up-regulated in the IRI-treated mice compared with the sham-operated mice (353.2±24.9 vs. 179.8±35.4 pg/mL, Figure 2C, Table S2). These data indicated that CLCF1 was up-regulated by IRI in the kidney.
FOXO3 negatively regulates CLCF1 expression
FOXO3 is a well-known transcription factor that negatively regulates the production of many inflammatory factors (15). To investigate whether CLCF1 expression can be regulated by FOXO3, we detected the circulatory CLCF1 levels in FOXO3-knockout mice. We observed that FOXO3 knockout did not alter the serum Cr levels in both the sham-operated and IRI-treated mice (Figure 3A,3B, Table S2) but that FOXO3 knockout did increase serum CLCF1 levels in the sham-operated mice (Figure 3C, Table S2). However, FOXO3 knockout failed to alter the CLCF1 levels in IRI-treated mice (Figure 3C, Table S2). These data indicated that FOXO3 might negatively regulate the expression of CLCF1.

Discussion
In the present study, we identified CLCF1 as an upregulated factor in renal IRI that may be regulated by FOXO3. Through the overlap analysis, we speculate that CLCF1 is a “bridge” molecule linking renal IRI to kidney transplantation DGF. Our findings may provide novel insights into the mechanisms of IRI-induced DGF in kidney transplantation.
IRI, an inevitable event during kidney transplantation, is characterized by the restriction of blood supply to an organ followed by the restoration of blood flow and re-oxygenation (16). IRI mainly involves 2 distinct stages: ischemia and reperfusion. Either warm ischemia or cold ischemia can cause a reduction in the supply of oxygen and nutrients, resulting in the accumulation of lactic acid and intracellular acidosis induced by enhanced anaerobic glycolysis (17). Furthermore, the Na/K ATPase pump is damaged, and the balance between intracellular and extracellular ions is destroyed (18). Cell death can be induced in the ischemia stage, and inflammatory or oxidative injury can be exacerbated during the reperfusion stage (19). IRI induces leukocyte occlusion in the capillaries (20), resulting in local inflammation of the microvasculature. These changes enhance the release of reactive oxygen species, and thus promote the production of inflammatory mediators and proteolytic enzymes (21). All these IRI mechanisms eventually result in renal damage, which may participate in the occurrence of DGF.
CLCF1 has been identified as an unreported upregulated gene. The upregulation of CLCF1 in both the IRI model database and the DGF database suggests that CLCF1 may play an important role in IRI-induced DGF after renal transplantation. It is worth mentioning that the upregulation of CLCF1 in the DGF database published by Mueller is most likely due to intrinsic factors of the donor kidneys and short-term acute kidney injury, while the upregulation of CLCF1 found in our experiments is due to acute kidney injury caused by IRI. This leads us to speculate that it may be the cumulative effect of the upregulation of CLCF1 in the donor kidney genome and the upregulation of CLCF1 caused by IRI which ultimately participates in the occurrence of DGF.
CLCF1 is a member of IL-6 family which is broadly expressed in secondary lymphoid organs such as lymph nodes and the spleen, as well as many other organs (bone marrow, ovary, placenta, kidney, pituitary gland). The secretion of CLCF1 is required to interact with CRLF1 or sCNTFR to form a heterodimer, which also presents as a circulatory form (12). The CLCF1/CRLF1 or CLCF1/sCNTFR heterodimer binds to the cell surface CNTFR and recruits gp130 and leukemia inhibitory factor receptor (LIFR), mediating the activation of STAT3 and the extracellular signal-regulated kinase 1/2/mitogen-activated protein kinase (ERK1/2/MAPK) pathways (22). Pro-inflammatory cytokines, such as tumor necrosis factor (TNF)-α, interferon (INF)-γ, and IL-6, can up-regulate the CRLF expression and thus enhance the action of CLCF1 (23). Therefore, increased levels of pro-inflammatory factors during IRI may contribute to an increase of CLCF1 levels in both the kidney tissues and sera. Mutations of CLCF1 or CRLF1 in humans lead to CISS (24), whereas CLCF1-deficient mice exhibit a severe suckling defect and cannot survive during the perinatal period, indicating that CLCF1 may be a crucial factor in the process of development (8,10). CLCF1 also supports the survival of embryonic motor and sympathetic neurons and may induce astrocyte differentiation of fetal neuroepithelial cells (25).
The function of CLCF1 remains far from being understood. Recent studies have indicated that CLCF1 attend to modulate mesenchymal stem cell osteoblastic differentiation (26) and promote macrophage-foam cell transition (27). For kidney diseases, CLCF1 has been reported to trigger the recurrence of FSGS after kidney transplantation (12). This effect is mainly associated with the activation of JAK2/STAT3, and inhibition of this signalling pathway can overcome the effect of CLCF1 on FSGS (12). In the present study, we observed that CLCF1 was significantly up-regulated in the kidney tissues of IRI-treated mice. These results indicate that CLCF1 might be a prominent biomarker of IRI-induced DGF.
A recent study reported that very-low-density lipoproteins could modulate CLCF1 activity (9). However, the regulation of CLCF1 expression is unclear. It is known that IL-6 is regulated by FOXO3. As both IL-6 and CLCF1 belong to the IL-6 family of cytokines, it can be speculated that CLCF1 may also be regulated by FOXO3. In the present study, we found that circulatory CLCF1 was significantly upregulated in FOXO3 knockout mice, indicating that FOXO3 might participate in the regulation of CLCF1 expression.
FOXO3 is a known transcription factor that is broadly expresses in many tissues and acts as an inflammation inhibitor (28). Loss of FOXO3 results in enhanced inflammation characterized by increased IL-6 levels (28). Our data indicated that circulatory CLCF1 was upregulated in FOXO3-knockout mice, indicating that FOXO3 may negatively regulate CLCF1 expression. However, whether FOXO3 inhibits CLCF1 expression directly or indirectly via additional mechanisms is yet to be understood. Our data indicated that the circulatory CLCF1 levels were comparable between wild type and FOXO3 knockout mice with renal IRI. However, we did not knock out CLCF1 in the mouse models because a deficiency of CLCF1 is embryonically lethal (8,10). CLCF1 is known to activate the JAK2/STAT3 pathway (12), we postulate that CLCF1 might participate in the regulation of renal IRI through this signaling pathway. However, these effects should be explored in future studies.
Medical products that limit the short-term deleterious effects of IRI or DGF and improve long-term allograft survival are urgently needed (29). Due to the high complexity of DGF, there is a lack of effective diagnosis and treatment methods for this condition. In addition, until recently, there have been no United States Food and Drug Administration (FDA)-approved treatments for DGF available (29). This study may provide evidence for using CLCF1 in diagnostic and therapeutic applications and promote further understanding of IRI-induced DGF.
In conclusion, CLCF1 is a potential biomarker of IRI-induced DGF. This finding may help to understand the link between IRI and DGF. We look forward to discovering more about CLCF1 in kidney transplantation in future studies.
Acknowledgments
We would like to thank Lisa-Jane Roberts and Calina Betlazar-Maseh for their help in polishing our paper.
Funding: This study was supported by the National Natural Science Foundation of China (No. 81870509).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-4381/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-4381/dss
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-4381/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-4381/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The animal experiments in this study were approved by the Beijing Friendship Hospital Animal Care and Use Committee (No. 18-2022). All procedures involving animals were performed and monitored in compliance with the guidelines of Beijing Friendship Hospital Animal Use Regulations.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chaumont M, Racapé J, Broeders N, et al. Delayed Graft Function in Kidney Transplants: Time Evolution, Role of Acute Rejection, Risk Factors, and Impact on Patient and Graft Outcome. J Transplant 2015;2015:163757. [Crossref] [PubMed]
- Iñiguez M, Berasain C, Martinez-Ansó E, et al. Cardiotrophin-1 defends the liver against ischemia-reperfusion injury and mediates the protective effect of ischemic preconditioning. J Exp Med 2006;203:2809-15. [Crossref] [PubMed]
- Ponticelli C. Ischaemia-reperfusion injury: a major protagonist in kidney transplantation. Nephrol Dial Transplant 2014;29:1134-40. [Crossref] [PubMed]
- Qiu L, Lai X, Wang JJ, et al. Kidney-intrinsic factors determine the severity of ischemia/reperfusion injury in a mouse model of delayed graft function. Kidney Int 2020;98:1489-501. [Crossref] [PubMed]
- Hu X, Su M, Lin J, et al. Corin Is Downregulated in Renal Ischemia/Reperfusion Injury and Is Associated with Delayed Graft Function after Kidney Transplantation. Dis Markers 2019;2019:9429323. [Crossref] [PubMed]
- Su M, Hu X, Lin J, et al. Identification of Candidate Genes Involved in Renal Ischemia/Reperfusion Injury. DNA Cell Biol 2019;38:256-62. [Crossref] [PubMed]
- Mueller TF, Reeve J, Jhangri GS, et al. The transcriptome of the implant biopsy identifies donor kidneys at increased risk of delayed graft function. Am J Transplant 2008;8:78-85. [PubMed]
- Sims NA. Cardiotrophin-like cytokine factor 1 (CLCF1) and neuropoietin (NP) signalling and their roles in development, adulthood, cancer and degenerative disorders. Cytokine Growth Factor Rev 2015;26:517-22. [Crossref] [PubMed]
- Pasquin S, Chehboun S, Dejda A, et al. Effect of human very low-density lipoproteins on cardiotrophin-like cytokine factor 1 (CLCF1) activity. Sci Rep 2018;8:3990. [Crossref] [PubMed]
- Hahn AF, Waaler PE, Kvistad PH, et al. Cold-induced sweating syndrome: CISS1 and CISS2: manifestations from infancy to adulthood. Four new cases. J Neurol Sci 2010;293:68-75. [Crossref] [PubMed]
- Zamanian JL, Xu L, Foo LC, et al. Genomic analysis of reactive astrogliosis. J Neurosci 2012;32:6391-410. [Crossref] [PubMed]
- Sharma M, Zhou J, Gauchat JF, et al. Janus kinase 2/signal transducer and activator of transcription 3 inhibitors attenuate the effect of cardiotrophin-like cytokine factor 1 and human focal segmental glomerulosclerosis serum on glomerular filtration barrier. Transl Res 2015;166:384-98. [Crossref] [PubMed]
- Dejean AS, Beisner DR, Ch'en IL, et al. Transcription factor Foxo3 controls the magnitude of T cell immune responses by modulating the function of dendritic cells. Nat Immunol 2009;10:504-13. [Crossref] [PubMed]
- Qi H, Tian D, Li M, et al. Foxo3 Promotes the Differentiation and Function of Follicular Helper T Cells. Cell Rep 2020;31:107621. [Crossref] [PubMed]
- Hwang JW, Rajendrasozhan S, Yao H, et al. FOXO3 deficiency leads to increased susceptibility to cigarette smoke-induced inflammation, airspace enlargement, and chronic obstructive pulmonary disease. J Immunol 2011;187:987-98. [Crossref] [PubMed]
- Malek M, Nematbakhsh M. Renal ischemia/reperfusion injury; from pathophysiology to treatment. J Renal Inj Prev 2015;4:20-7. [PubMed]
- Han JY, Horie Y, Fan JY, et al. Potential of 3,4-dihydroxy-phenyl lactic acid for ameliorating ischemia-reperfusion-induced microvascular disturbance in rat mesentery. Am J Physiol Gastrointest Liver Physiol 2009;296:G36-44. [Crossref] [PubMed]
- Alejandro VS, Nelson WJ, Huie P, et al. Postischemic injury, delayed function and Na+/K(+)-ATPase distribution in the transplanted kidney. Kidney Int 1995;48:1308-15. [Crossref] [PubMed]
- Zheng J, Wei CC, Hase N, et al. Chymase mediates injury and mitochondrial damage in cardiomyocytes during acute ischemia/reperfusion in the dog. PLoS One 2014;9:e94732. [Crossref] [PubMed]
- Sabido F, Milazzo VJ, Hobson RW 2nd, et al. Skeletal muscle ischemia-reperfusion injury: a review of endothelial cell-leukocyte interactions. J Invest Surg 1994;7:39-47. [Crossref] [PubMed]
- Abdallah DM, Nassar NN, Abd-El-Salam RM. Glibenclamide ameliorates ischemia-reperfusion injury via modulating oxidative stress and inflammatory mediators in the rat hippocampus. Brain Res 2011;1385:257-62. [Crossref] [PubMed]
- Vicent S, Sayles LC, Vaka D, et al. Cross-species functional analysis of cancer-associated fibroblasts identifies a critical role for CLCF1 and IL-6 in non-small cell lung cancer in vivo. Cancer Res 2012;72:5744-56. [Crossref] [PubMed]
- Rebane A, Runnel T, Aab A, et al. MicroRNA-146a alleviates chronic skin inflammation in atopic dermatitis through suppression of innate immune responses in keratinocytes. J Allergy Clin Immunol 2014;134:836-847.e11. [Crossref] [PubMed]
- Tüysüz B, Kasapçopur O, Yalçınkaya C, et al. Multiple small hyperintense lesions in the subcortical white matter on cranial MR images in two Turkish brothers with cold-induced sweating syndrome caused by a novel missense mutation in the CRLF1 gene. Brain Dev 2013;35:596-601. [Crossref] [PubMed]
- Uemura A, Takizawa T, Ochiai W, et al. Cardiotrophin-like cytokine induces astrocyte differentiation of fetal neuroepithelial cells via activation of STAT3. Cytokine 2002;18:1-7. [Crossref] [PubMed]
- Nahlé S, Pasquin S, Laplante V, et al. Cardiotrophin-like cytokine (CLCF1) modulates mesenchymal stem cell osteoblastic differentiation. J Biol Chem 2019;294:11952-9. [Crossref] [PubMed]
- Pasquin S, Laplante V, Kouadri S, et al. Cardiotrophin-like Cytokine Increases Macrophage-Foam Cell Transition. J Immunol 2018;201:2462-71. [Crossref] [PubMed]
- Viatte S, Lee JC, Fu B, et al. Association Between Genetic Variation in FOXO3 and Reductions in Inflammation and Disease Activity in Inflammatory Polyarthritis. Arthritis Rheumatol 2016;68:2629-36. [Crossref] [PubMed]
- Cavaillé-Coll M, Bala S, Velidedeoglu E, et al. Summary of FDA workshop on ischemia reperfusion injury in kidney transplantation. Am J Transplant 2013;13:1134-48. [Crossref] [PubMed]


