
Downregulation of lncRNA NEAT1 inhibits the proliferation of human cutaneous squamous cell carcinoma in vivo and in vitro
Introduction
An increase in the incidence of skin cancer, a malignant tumor, has been observed in recent years (1). Skin cancer is the general term used to refer to malignant tumors of the skin, which include cutaneous squamous cell carcinoma (CSCC), melanoma, and basal cell carcinoma. Among them, basal cell carcinoma and CSCC are the most common (2). Melanoma accounts for only 2% of the total number of cases of cutaneous carcinoma, but due to its high degree of malignancy and strong invasiveness, it has the highest mortality (3). About 95% of all skin cancer cases are caused by squamous cell carcinoma. Since squamous cell carcinoma has no typical early symptoms, it is difficult to diagnose, and as a result, most patients have developed invasive disease by the time they are first diagnosed, which greatly limits the treatment (4). Approximately 5% of patients with CSCC have lymph node metastasis at the time of initial diagnosis (2,5). In some cases, CSCC may have metastasized to the viscera due to its rapid progression. Fewer than 20% of CSCC patients with localized or distant lymphatic metastasis survive 10 years after their diagnosis, while patients with distant metastasis have a 10-year survival rate of 10% or less (6), illustrating the significant challenge in treating patients with advanced and metastatic squamous cell carcinoma.
Since the development of next-generation sequencing technology in recent years, mutations of related genes have been discovered within squamous cell carcinoma tissues, providing researchers with a reference for the prevention, diagnosis, and treatment of tumors. Despite this, squamous cell carcinoma mortality rates remain steadily on the rise (7). It is crucial that key signal molecules or diagnostic markers that regulate the proliferation of squamous cell carcinoma are detected in order to hinder tumor invasion and metastasis as well as improve radiotherapy effectiveness.
Long noncoding RNAs (lncRNAs) are species- and tissue-specific and have a length of more than 200 nt. LncRNAs can interact with deoxyribonucleic acid (DNA), RNAs, and proteins, and they regulate a wide range of biological processes (8,9). The lncRNA nuclear-enriched abundant transcript 1 (NEAT1) has been implicated in the growth and metastasis of esophageal squamous cell carcinoma (ESCC), colorectal cancer, and neck and head squamous cell carcinoma (10-12). Ma et al. (13) found that lncRNA NEAT1 regulated toxic T cell infiltration in lung cancer by inhibiting the cyclic GMP-AMP synthase (cGAS)/stimulator of interferon genes (STING) pathway. Zhang et al. (10) reported that lncRNA NEAT1 activated the Wnt signaling pathway to promote the progression and metastasis of colorectal cancer. Kou et al. (14) found that lncRNA NEAT1 expression was increased in liver cancer tissues, and downregulation of lncRNA NEAT1 inhibited epidermal growth factor receptor (EGFR) expression, promoted apoptosis of liver cancer cells, and inhibited the cell cycle, thereby impeding tumor proliferation and invasion. These studies indicate that lncRNA NEAT1 is highly expressed in many malignant tumors and thus may have good potential for application in the early diagnosis, treatment, and prognosis of various cancers. At present, no reports have been published concerning the role of lncRNA NEAT1 in the genesis and development of squamous cell carcinoma. This study is the first to investigate the role of lncRNA NEAT1 in the pathogenesis and development of CSCC. We detected the expression level of lncRNA NEAT1 in CSCC tissues and analyzed the effects of downregulated lncRNA NEAT1 expression on cell proliferation, colony formation, invasion, migration, and preliminary molecular mechanisms. In addition, in vivo experiments confirmed that knockdown of lncRNA NEAT1 affected the proliferation of CSCC cells. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6916/rc).
Methods
Clinical sample collection
The study involving human experiments was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Those who took part in the study provided informed consent and the study received approval from the ethics board of The First Affiliated Hospital of Soochow University (No. 2018KT124).
A total of 54 pairs of CSCC tissues and paracancer tissues were collected from CSCC patients at The First Affiliated Hospital of Soochow University between January 2018 and February 2019. No preoperative treatment was given. Cancer and adjacent tissues removed during surgery were collected, rinsed with phosphate buffer saline (PBS), and then stored in liquid nitrogen for 30 minutes. Reverse transcription was used to obtain complementary DNA (cDNA) from tissue messenger RNA (mRNA). In addition, we collected relevant clinical data from patients.
Cell culture
CSCC cell lines (A431, SCL-1, SCC13, and HSC-5) and human normal skin cell line (HaCaT) were procured from the Shanghai Institutes for Biological Sciences (Shanghai, China). The cells were grown in Roswell Park Memorial Institute (RPMI)-1640 medium containing 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA) and antibiotics (streptomycin, 100 µg/mL; penicillin, 100 U/mL) in a 37 °C incubator containing CO2 (5%).
Reverse transcription polymerase chain reaction (RT-PCR) assay
Extraction of total RNA from sample cells and tissues was done using Trizol reagent (Invitrogen, Carlsbad, CA, USA). NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA) was used to determine the concentration of total RNA. Reverse transcription of lncRNA NEAT1 was performed using a reverse transcription kit from Invitrogen. CDNA was synthesized by reverse transcription. The conditions for reverse transcription were 42 °C 30 minutes, 85 °C 5 minutes. CDNA amplification was performed using fluorescence quantitative PCR (qPCR; Thermo Fisher Scientific). The expression of lncRNA NEAT1 was detected using Applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fisher Scientific). The double standard curve method was used to determine the relative quantitative value of each gene. In our analysis, we used the 2−ΔΔcycle threshold (Ct) method, in which ΔCt = target gene Ct value − internal reference gene Ct value, and ΔΔCt = transfection group ΔCt − control group ΔCt.
lncRNA NEAT1:
Forward: 5'-TTGGGACAGTFFACGTGTGG-3';
Reverse: 5'-TCAAGTCCAGCAGAGCA-3';
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH):
Forward: 5'-TGACGTGCCGCCTGGAGAAC-3';
Reverse: 5'-CCGGCATCGAAGGTGGAAGAG-3'.
Cell transfection
A plasmid U6/GFP/Neo with short hairpin RNA (shRNA) targeting lncRNA NEAT1 was constructed, synthesized, and cloned with negative control and shRNA. The transfection was performed with Lipofectamine 3000 (Merck, Kenilworth, NJ, USA). Cells in the logarithmic growth stage first digested by trypsin and inoculated into 6-well plates. We then gently shook the plates to ensure the even distribution of the cells. Approximately 12 hours after achieving 85–90% cell density, transfection was performed. The medium was replaced with a fresh basal medium with no serum and antibiotics 2 hours prior to transfection. Transfection was performed by mixing 0.2 µg of plasmid, 0.4 µL of Lipofectamine 3000, and 5 µL of Opti-Minimal Essential Medium (MEM, Invitrogen) medium in 6-well plates.
NEAT1 shRNA1: 5'-CACCTGTTTGCCTGCCTTCTT-3';
NEAT1 shRNA1: 5'-ACGCAGCAGATCAGCATCCTT-3';
Scramble for shRNA: 5'-TTCTCCGAACGTGTCACGT-3'.
MTT assay
The proliferative capacity of cells was measured using 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide (MTT). Cells in the logarithmic growth stage were digested by trypsin and inoculated in 96-well plates (3×103 cells/well). At specific time intervals, MTT (0.5 mg/mL, Gibco, Waltham, MA, USA) was added to the medium. The plates were incubated for 4 hours at 37 °C, and the supernatants were discarded. We then added dimethyl sulfoxide (150 µL) to each well and measured the optical density (OD) on a microplate reader (Thermo Fisher Scientific).
Colony formation assay
A colony formation assay was performed to test the colony-forming capacity of cells. Trypsin digestion was used to digest cells in the logarithmic growth stage, and 600 cells/well were inoculated into 6-well plates. Cells were grown in an incubator with CO2 (5%) at 37 °C, and the growth medium was changed every 3 days. We cultured the cells for 21 days, fixed them in 75% methanol for 10 minutes, washed them 3 times in PBS, followed by crystal violet (0.1%) staining for 15 minutes. Using a microscope and a camera, the cells were counted and photographed.
Invasion and migration assays
Transwell assay was used to measure the invasive and migratory capacity of cells. Cells in the logarithmic growth stage were first digested by trypsin, centrifuged, and suspended in a medium containing 1% serum. The cells were then seeded in the upper chamber of 24-well plates (BD Biosciences, Franklin Lakes, NJ, USA) at a density of 2×104 cells/well. A medium containing 15% serum was added to the lower chamber and incubated at 37 °C with 5% CO2 for 36 hours. Nonmigrated cells were gently removed from the upper chamber using cotton swabs. After being fixed with 95% ethanol and 5% acetic acid for 30 minutes, the cells were stained with crystal violet (0.1%) for 15 minutes and then photographed and counted under a TE2000-U inverted microscope (Nikon, Tokyo, Japan). Invasion assays were conducted by treating the upper chamber of 24-well plates with Matrigel (20 µg/mg, BD Biosciences). The plates were then incubated for 1 hour at 37 °C, followed by an invasion assay procedure.
Western blot
A protein extraction kit (Abcam, Shanghai, China) was used to isolate the total protein from cells in the logarithmic growth phase. In addition to quantification with a bicinchoninic acid (BCA) protein quantification kit (Abcam), the total protein was electrophoresed with 8–10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE; Shanghai Protophylla, China), transferred to a polyvinylidene fluoride (PVDF) membrane, and blocked with skim milk (5%) at room temperature for 2 hours. The PVDF membrane was then incubated for 12 hours at 4 °C with a primary antibody. The primary antibodies used were E-cadherin (1:800; ab76055), N-cadherin (1:800; ab211126), matrix metalloproteinase 2 (MMP-2; 1:1,000, ab37150), vimentin (1:1,000; ab24525), MMP-9 (1:1,000; ab73734), and GAPDH (1:1,000; AB8245). After 2 hours of incubation with a secondary antibody, the membrane was washed 5 times in PBS buffer, and the developer was added to detect protein signals.
Subcutaneous tumorigenesis of nude mice in vivo
A group of 10 6-week-old BALB/c nude mice (weighing 15–18 g) were obtained from Nanjing Model Animal Center and grown under specific pathogen-free conditions. A cell density of 107/mL was adjusted after collecting the transfected CSCC cells. A 0.1 mL suspension was subcutaneously injected into the nude mice, and the tumor-bearing tissues were measured and observed after 10 days. The length and width of the tumor-bearing tissues were measured every 3 days. The volume of the transplanted tumor was determined using the following formula: V = [(a×b×b)/2]; V: the volume of the transplant, a: the long diameter, and b: the short diameter of the tumor. To determine the tumor volume and weight, the mice were sacrificed after 28 days, and the total mRNA was extracted from the tumor-bearing tissue. Fluorescence qPCR was used to detect lncRNA NEAT1 expression in the tumor-bearing tissue.
Experiments involving animals were performed under a project license (No. S20200314-034) granted by the ethics board of The First Affiliated Hospital of Soochow University, in compliance with Chinese guidelines for the care and use of animals.
Statistical analysis
The data were analyzed using SPSS v19.0. Measurement data were presented as mean ± standard deviation. When data conformed to a normal distribution, the t-test or analysis of variance was used for inter-group comparisons, while the nonparametric rank sum test was used for nonnormally distributed measurement data. Data were counted using the Chi-square test. P<0.05 was considered significant.
Results
LncRNA NEAT1 is highly expressed in CSCC tissues and is associated with patients’ clinical and pathological features
LncRNA NEAT1 expression was detected in 54 pairs of CSCC and paracancer tissues using fluorescence qPCR. In contrast to paracancer tissue, lncRNA NEAT1 was highly expressed in CSCC tissues (Figure 1A). On the basis of lncRNA NEAT1 expression levels, CSCC tissues were assigned to a high (>3.67, n=35) or low (≤3.67, n=19) NEAT1 lncRNA expression group, and the correlation between NEAT1 lncRNA expression level and clinicopathological features of patients was analyzed. The results demonstrated a correlation between high NEAT1 expression, lymph node metastasis, and the tumor, regional lymph nodes, and distant metastasis (TNM) grade (Table 1). Additionally, NEAT1 was detected by fluorescent qPCR in CSCC cell lines (SCC13, A431, SCL-1, and HSC-5) and human normal skin cell line (HaCaT) (Figure 1B). Compared with HaCaT, lncRNA NEAT1 was highly expressed in CSCC cells, with the highest expression in A431 cells and HSC-5 cells. These results suggested that lncRNA NEAT1 could be used to diagnose CSCC.
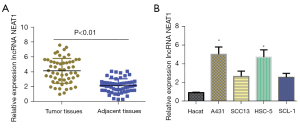
Table 1
| Characteristics | Number (n=54) | Low (n=19) | High (n=34) | P value |
|---|---|---|---|---|
| Age (years) | 0.9794 | |||
| ≤60 | 31 | 11 | 19 | |
| >60 | 23 | 9 | 15 | |
| Gender | 0.1956 | |||
| Male | 26 | 7 | 21 | |
| Female | 28 | 12 | 13 | |
| Tumor size (cm) | 0.1213 | |||
| ≤4 | 32 | 9 | 13 | |
| >4 | 22 | 10 | 22 | |
| Lymphatic metastasis | 0.0224* | |||
| Yes | 25 | 8 | 25 | |
| No | 29 | 11 | 9 | |
| Differentiation | 0.5945 | |||
| Well | 10 | 4 | 11 | |
| Moderate | 19 | 5 | 9 | |
| Poor | 25 | 10 | 14 | |
| TNM | 0.0111* | |||
| I & II | 21 | 15 | 6 | |
| III & IV | 42 | 17 | 25 |
*, Chi-square test was used for the association analysis and P<0.05 was deemed statistically significant. lncRNA, long noncoding RNA; NEAT1, nuclear-enriched abundant transcript 1; TNM, tumor, node, metastasis.
Knockdown of lncRNA NEAT1 impedes the proliferation of CSCC
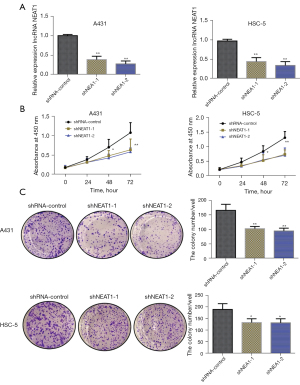
The above results showed that NEAT1 was highly expressed in A431 and HSC-5 cells. To further study the role of lncRNA NEAT1 in the occurrence and development of CSCC, 2 lentiviral vectors targeting the NEAT1 sequence, short hairpin of NEAT1 (shNEAT1)-1 and shNEAT1-2 as well as a blank control cell line (shNAT1-control), were created (Figure 2A). The knockdown efficiency of lncRNA NEAT1 in CSCC was determined using fluorescence qPCR. The results showed that transfection with shNEAT1-1 and shNEAT1-2 interference sequences dramatically knocked down lncRNA NEAT1 expression in A431 and HSC-5 cells compared with the blank control group (shNAT1-control) (Figure 2B). As shown in the MTT assay results, NEAT1 knockdown significantly reduced the proliferation ability of A431 and HSC-5 cells compared with the blank control group (shNAT1-control group). Based on the results of our plate colony formation assay, lncRNA NEAT1 knockdown markedly inhibited A431 and HSC-5 colony formation compared with the control group (shNAT1-control group) (Figure 2C). These results showed that the knockdown of lncRNA NEAT1 significantly inhibited the proliferation of CSCC.
NEAT1 lncRNA knockdown inhibits epithelial-mesenchymal transition (EMT)-related protein expression in CSCC
Tumor cells are characterized by their ability to invade and migrate. We examined the effect of lncRNA NEAT1 on the invasion and migration abilities of CSCC cells using a Transwell assay. The results showed that A431 and HSC-5 cells had significantly diminished invasive and migrating abilities when lncRNA NEAT1 was knocked down compared with the control group (shNAT1-control group) (Figure 3).

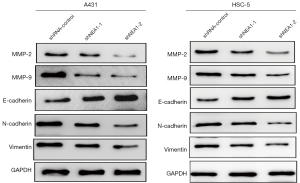
A western blot assay was conducted to determine the outcome of knockdown of lncRNA NEAT1 on the expression of key proteins involved in the invasion and migration of CSCC cells in order to further investigate the molecular mechanism of its inhibition. The results of lncRNA NEAT1 knockdown revealed that it significantly reduced matrix metalloproteinase-2 and -9 expression in CSCC cells, A431, and HSC-5 (Figure 4). Moreover, lncRNA NEAT1 knockdown significantly affected the level of proteins related to EMT, including enhancing of E-cadherin expression and inhibiting the expression of N-cadherin and vimentin. These results showed that lncRNA NEAT1 knockdown could significantly modify invasion and migration-related proteins in CSCC cells.
Loss of NEAT1 lncRNA expression impedes CSCC proliferation in vivo
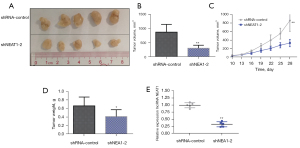
The above in vitro assays indicated that loss of NEAT1 lncRNA significantly impeded proliferative and invasive abilities in CSCC cells. We then used nude mice to investigate whether further knockdown of lncRNA NEAT1 could affect CSCC proliferation in vivo. After 28 days, the results showed that lncRNA NEAT1 knockdown significantly inhibited CSCC proliferation in vivo (Figure 5A,5B). The average weight and volume of the tumor-bearing tissues in the shNEAT-2 group were remarkably reduced compared with the blank control group (shNAT1-control) (Figure 5C,5D). RNA was extracted from tumors, and fluorescence qPCR was used to measure NEAT1 expression. A significant reduction in expression of NEAT1 lncRNA was observed in the shNEAT1-2 group relative to the blank control group (shNAT1-control) (Figure 5E), suggesting that knockdown of NEAT1 lncRNA in nude mice could significantly attenuate CSCC proliferation.
Discussion
Several studies have shown that long-neglected RNAs with no coding ability, such as lncRNAs, play an important role in squamous cell carcinoma genesis and progression (15-17). According to recent studies (10,18), lncRNA NEAT1 is strongly related to tumor proliferation, metastasis, and radiotherapy tolerance. To date, no studies have examined the correlation between NEAT1 lncRNA and squamous cell carcinoma. In the present study, RNA sequencing confirmed that lncRNA NEAT1 was expressed in squamous cell carcinoma tissues and correlated with both TNM grade and lymph node metastasis in patients. The knockdown of NEAT1 in CSCC cells impaired proliferation, metastasis, and invasion in both in vitro and in vitro experiments. Based on the results of the molecular mechanism study, knockdown of lncRNA NEAT1 significantly inhibited metalloproteinase-2, and -9, N-cadherin, and vimentin expression and promoted the expression of E-cadherin.
CSCC, which originates from keratinocytes, is the second most common form of nonmelanoma skin cancer and one of the primary causes of nonmelanoma skin cancer-related death (19). Overexpression of some lncRNAs has been found in a number of tumor tissues and thus can be considered potential biomarkers or targets for future diagnosis and treatment of squamous cell carcinoma (17). For example, LINC00162 has been found to be overexpressed in squamous cell carcinoma cells, with knockdown of LINC00162 impeding the migration and proliferation of squamous carcinoma cells and inhibiting the in vivo growth of human squamous cell carcinoma xenografts. Further, it has been reported that knockdown of LINC00162 inhibits squamous cell carcinoma cells’ upregulation of extracellular signal-regulated kinase 1/2 (ESRK1/2) activity and expression of dual-specificity phosphatase 6 (DUSP6) (16). Thus, the identification of abnormally expressed lncRNAs in squamous cell carcinoma tissues and detailed investigation of their biological functions may shed light on the molecular regulation of squamous cell carcinoma at the level of noncoding RNA, thereby providing a basis for early diagnosis and targeted treatment of squamous cell carcinoma. This study found that lncRNA NEAT1 expression was highly correlated with TNM stage and metastasis of lymph nodes in patients with squamous cell carcinoma, suggesting that lncRNA NEAT1 expression levels could be used as a potential indicator for CSCC pathological grading.
LncRNA NEAT1 regulates a variety of physiological functions, including regulation of signaling pathways such as toll-like receptor 2 (TLR2) and nuclear factor kappa-B in response to septicemia-induced myocardial injury (20). A reduction in the protein level of signal transducer and activator of transcription 3 is also observed when NEAT1 is inhibited (21). Furthermore, lncRNA NEAT1 plays a role in regulating choroidal neovascularization and M2 macrophage polarization (22). Cancer cells from the endometrium and breast may also be significantly slowed by lncRNA NEAT1 (23,24). Results from previous studies have shown that knockdown of lncRNA NEAT1 could significantly inhibit the proliferation and colony formation of squamous cell carcinoma. Tumor cells have been shown to promote metastasis to distant sites by increasing their invasive and migrating abilities, which is the cause of cancer recurrence (25). MMP-2 and MMP-9 genes play a significant part in squamous cell carcinoma metastasis (26). For example, microRNA (miR)-766 promotes the invasion and migration of squamous cell carcinoma cells A431 and SCL-1 by releasing MMP-2 and -9 (27). In CSCC cells, overexpression of LINC00520 significantly inhibits migratory and adhesive capacity by downregulating MMP-2 mRNA and MMP-9 protein levels (28). A subsequent investigation in this study revealed that the knockdown of lncRNA NEAT1 remarkably inhibited the migratory and invasive capacity of squamous cell carcinoma. According to the results of molecular mechanism studies, the knockdown of NEAT1 significantly inhibited the expression of metalloproteinase-2, -9, vimentin, and N-cadherin and promoted the expression of E-cadherin. However, it is not known whether lncRNA NEAT1 regulates the expression of the above molecules directly or indirectly. Numerous scientific studies have shown that lncRNA regulates cell function in 3 ways: (I) binding to gene targets, thereby elevating or lowering target genes; (II) modifying histone structures or increasing transcription factors; and (III) interacting with miRNA to affect gene expression (29-31). We will continue to study the molecular mechanisms regulating lncRNA NEAT1 in CSCC in future studies.
Acknowledgments
Funding: The study was supported by the Medical Research Project of Jiangsu Provincial Health Commission (Z2019031), the Medical Research Project of Nantong Health Commission (MA2020001), and the Nantong Science and Technology Bureau (MS12021093, JCZ21126).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6916/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6916/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6916/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study involving human experiments was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and approved by the ethics board of The First Affiliated Hospital of Soochow University (No. 2018KT124). Informed consent was taken from all individual participants. Experiments involving animals were performed under a project license (No. S20200314-034) granted by the ethics board of The First Affiliated Hospital of Soochow University, in compliance with Chinese guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Schierbeck J, Vestergaard T, Bygum A. Skin Cancer Associated Genodermatoses: A Literature Review. Acta Derm Venereol 2019;99:360-9. [Crossref] [PubMed]
- Waldman A, Schmults C. Cutaneous Squamous Cell Carcinoma. Hematol Oncol Clin North Am 2019;33:1-12. [Crossref] [PubMed]
- Ahmed B, Qadir MI, Ghafoor S. Malignant Melanoma: Skin Cancer-Diagnosis, Prevention, and Treatment. Crit Rev Eukaryot Gene Expr 2020;30:291-7. [Crossref] [PubMed]
- Que SKT, Zwald FO, Schmults CD. Cutaneous squamous cell carcinoma: Incidence, risk factors, diagnosis, and staging. J Am Acad Dermatol 2018;78:237-47. [Crossref] [PubMed]
- Brunssen A, Waldmann A, Eisemann N, et al. Impact of skin cancer screening and secondary prevention campaigns on skin cancer incidence and mortality: A systematic review. J Am Acad Dermatol 2017;76:129-139.e10. [Crossref] [PubMed]
- Rosenberg AR, Tabacchi M, Ngo KH, et al. Skin cancer precursor immunotherapy for squamous cell carcinoma prevention. JCI Insight 2019;4:e125476. [Crossref] [PubMed]
- Stratigos AJ, Garbe C, Dessinioti C, et al. European interdisciplinary guideline on invasive squamous cell carcinoma of the skin: Part 1. epidemiology, diagnostics and prevention. Eur J Cancer 2020;128:60-82. [Crossref] [PubMed]
- Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017;36:5661-7. [Crossref] [PubMed]
- Zhang J, Guo S, Piao HY, et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem 2019;75:379-89. [Crossref] [PubMed]
- Zhang M, Weng W, Zhang Q, et al. The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol 2018;11:113. [Crossref] [PubMed]
- Kozłowska J, Kozioł K, Stasiak M, et al. The role of NEAT1 lncRNA in squamous cell carcinoma of the head and neck is still difficult to define. Contemp Oncol (Pozn) 2020;24:96-105. [Crossref] [PubMed]
- Jiang Y, Cao W, Wu K, et al. LncRNA LINC00460 promotes EMT in head and neck squamous cell carcinoma by facilitating peroxiredoxin-1 into the nucleus. J Exp Clin Cancer Res 2019;38:365. [Crossref] [PubMed]
- Ma F, Lei YY, Ding MG, et al. LncRNA NEAT1 Interacted With DNMT1 to Regulate Malignant Phenotype of Cancer Cell and Cytotoxic T Cell Infiltration via Epigenetic Inhibition of p53, cGAS, and STING in Lung Cancer. Front Genet 2020;11:250. [Crossref] [PubMed]
- Kou JT, Ma J, Zhu JQ, et al. LncRNA NEAT1 regulates proliferation, apoptosis and invasion of liver cancer. Eur Rev Med Pharmacol Sci 2020;24:4152-60. [PubMed]
- Zhou W, Zhang S, Li J, et al. lncRNA TINCR participates in ALA-PDT-induced apoptosis and autophagy in cutaneous squamous cell carcinoma. J Cell Biochem 2019;120:13893-902. [Crossref] [PubMed]
- Piipponen M, Nissinen L, Farshchian M, et al. Long Noncoding RNA PICSAR Promotes Growth of Cutaneous Squamous Cell Carcinoma by Regulating ERK1/2 Activity. J Invest Dermatol 2016;136:1701-10. [Crossref] [PubMed]
- Wang Y, Sun B, Wen X, et al. The Roles of lncRNA in Cutaneous Squamous Cell Carcinoma. Front Oncol 2020;10:158. [Crossref] [PubMed]
- Gao Y, Fang P, Li WJ, et al. LncRNA NEAT1 sponges miR-214 to regulate M2 macrophage polarization by regulation of B7-H3 in multiple myeloma. Mol Immunol 2020;117:20-8. [Crossref] [PubMed]
- Lazar AD, Dinescu S, Costache M. Deciphering the Molecular Landscape of Cutaneous Squamous Cell Carcinoma for Better Diagnosis and Treatment. J Clin Med 2020;9:2228. [Crossref] [PubMed]
- Wang SM, Liu GQ, Xian HB, et al. LncRNA NEAT1 alleviates sepsis-induced myocardial injury by regulating the TLR2/NF-κB signaling pathway. Eur Rev Med Pharmacol Sci 2019;23:4898-907. [PubMed]
- Shui X, Chen S, Lin J, et al. Knockdown of lncRNA NEAT1 inhibits Th17/CD4+ T cell differentiation through reducing the STAT3 protein level. J Cell Physiol 2019;234:22477-84. [Crossref] [PubMed]
- Zhang P, Lu B, Zhang Q, et al. LncRNA NEAT1 Sponges MiRNA-148a-3p to Suppress Choroidal Neovascularization and M2 macrophage polarization. Mol Immunol 2020;127:212-22. [Crossref] [PubMed]
- Li X, Deng S, Pang X, et al. LncRNA NEAT1 Silenced miR-133b Promotes Migration and Invasion of Breast Cancer Cells. Int J Mol Sci 2019;20:3616. [Crossref] [PubMed]
- Wang W, Ge L, Xu XJ, et al. LncRNA NEAT1 promotes endometrial cancer cell proliferation, migration and invasion by regulating the miR-144-3p/EZH2 axis. Radiol Oncol 2019;53:434-42. [Crossref] [PubMed]
- Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol 2019;29:212-26. [Crossref] [PubMed]
- Zhang X, Wu L, Xiao T, et al. TRAF6 regulates EGF-induced cell transformation and cSCC malignant phenotype through CD147/EGFR. Oncogenesis 2018;7:17. [Crossref] [PubMed]
- Liu P, Shi L, Ding Y, et al. MicroRNA-766 Promotes The Proliferation, Migration And Invasion, And Inhibits The Apoptosis Of Cutaneous Squamous Cell Carcinoma Cells By Targeting PDCD5. Onco Targets Ther 2020;13:4099-110. [Crossref] [PubMed]
- Mei XL, Zhong S. Long noncoding RNA LINC00520 prevents the progression of cutaneous squamous cell carcinoma through the inactivation of the PI3K/Akt signaling pathway by downregulating EGFR. Chin Med J (Engl) 2019;132:454-65. [Crossref] [PubMed]
- Ali T, Grote P. Beyond the RNA-dependent function of LncRNA genes. Elife 2020;9:60583. [Crossref] [PubMed]
- Jathar S, Kumar V, Srivastava J, et al. Technological Developments in lncRNA Biology. Adv Exp Med Biol 2017;1008:283-323. [Crossref] [PubMed]
- Charles Richard JL, Eichhorn PJA. Platforms for Investigating LncRNA Functions. SLAS Technol 2018;23:493-506. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


