
IL-1B can serve as a healing process and is a critical regulator of diabetic foot ulcer
Introduction
Approximately 3.6 million people in Britain have been diagnosed with diabetes mellitus (DM), and it is estimated that this figure will increase to 5.0 million in the next 10 years (1). Despite the better management of these DM patients, the risk of DM complications also increases with time. Devastating macrovascular complications (e.g., cardiovascular disease) and microvascular complications (e.g., diabetic kidney disease, diabetic retinopathy, and neuropathy) increase the mortality of DM patients (1). Such complications may be correlated to gene alterations (1).
In Western countries, the annual incidence of foot ulceration in the diabetic population is around 2% (2). Diabetic foot (DF) is one of the most common end-point complications of diabetes (2). More than 2/3 of non-traumatic lower limb amputations are preceded by an ulcer (84%); thus, this pivotal event could enable early treatment (3). Among of the end-point complications, peripheral artery disease and neuropathy are the main causes of foot ulcers. These complications often act in combination, but can also act alone. DM patients with artery disease and neuropathy are asymptomatic in the early stages of DF, but as the disease develops, it causes chronic non-healing foot ulcers. Diabetic foot ulcers (DFUs) are a major medical and economic problem for health management agencies and clinical doctors globally. The regulators underlying DFU development and the healing process need to be urgently understood to enable the early diagnosis of DFU and the development of novel treatment methods.
Aside from the main etiological factors, several factors also have critical roles in the occurrence and processes of DFU, including abnormal gene alterations, immune cell infiltration, and inflammation (4-6). Wu et al. found that the downregulation of endothelium and lymphocyte associated ASCH domain 1 (EOLA1) expression exacerbated DFU by enhancing the inflammatory pathway activity, and also detected differentially expressed genes (DEGs), including metallothionein 2A and interleukin (IL)-6 (6).
Inflammation has been proven to be an important biological process in DFU (7,8). Various immune cells and cytokines drive the inflammation response. The M1 and M2 macrophage ratio was found to be more dysregulated in the skin of diabetic rabbits than control rabbits (7). Sawaya et al. found the FOXM1 inhibits immune cell infiltration in the DFU microenvironment and promotes the healing process (5). During the inflammation response, the activated inflammatory cells can produce several pro-inflammatory cytokines. Dinh et al. showed that unhealed DFU patients had higher expression levels of tumor necrosis factor-α, and matrix metallopeptidase 9 than in healed DFU patients (9). Thus, immune cell infiltration and the production of cytokines play a regulatory role in the DFU healing process. In this study, we sought to investigate differential gene expression, significant immune gene expression levels, and immune cell infiltration differences between healed and non-healed DFU patients. Through the study of these immune-related DEGs and the immune cells infiltration of healed DFU, we try to find the potential targets for the treatment of DFU.
We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-75/rc).
Methods
Raw data acquisition
The GSE143735 (comprising 4 common diabetes samples, 4 unhealed DFU samples, and 5 healed DFU samples) and GSE134431 (comprising 8 common diabetes samples, 6 unhealed DFU samples, and 7 healed DFU samples) transcriptome data sets were obtained from the Gene Expression Omnibus database. The transcriptome data were merged and normalized for further analysis. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
DEGs associated with DFU
To identify the ulcer-related DEGs in DFU, we first analyzed the differential expression between the samples with and without ulcers. We then analyzed the differential expression between the healed and unhealed samples to identify the healing-related DEGs in DFU.
Functional enrichment analysis of ulcer-related DEGs and healing-related DEGs in DFU
We performed gene ontology (GO) functional and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analyses of ulcer-related DEGs and healing-related DEGs through the clusterProfiler package in R, and used the ggplot2 package to present the results.
Intersection genes for ulcer-related DEGs and healing-related DEGs and immune-related genes
An immune-related gene list was downloaded from ImmPort. The intersection genes for the ulcer-related DEGs, healing-related DEGs, and immune-related genes were considered hub biomarkers and used for further analysis.
Immune infiltration analysis
We conducted a single-sample gene set enrichment analysis (ssGSEA) to analyze the immune landscapes of the DF patients. The ssGSEA is a machine learning algorithm that characterizes intratumoral immune landscapes and that can identify the expression of 28 kinds of immune cells. We compared the infiltration of 28 immune cells in common diabetes samples, unhealed DFU samples, and healed DFU samples. We also analyzed the relationship between the abundance of the 28 immune cells and the expression of immune-related hub biomarkers in DFU.
Statistical analysis
All of the statistical analyses were performed in R software. The difference analysis and correlation analysis were performed by Wilcoxon and Spearman correlations, respectively. A P value <0.05 was considered statistically significant.
Results
PCA of raw transcriptome data before and after data normalization
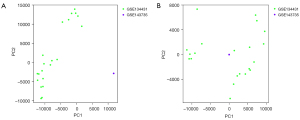
The principal component analysis (PCA) indicated that before normalization, the principal component of the gene expressions of the GSE143735 and GSE143431 data sets differed significantly (Figure 1A); however, the principal components of the gene expressions of these 2 data sets were at the same level after standardization (Figure 1B).
Ulcer-related DEGs and healing-related DEGs in DFU
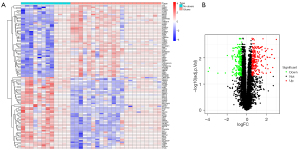
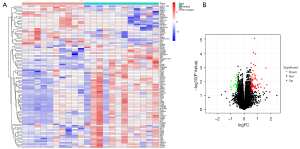
The ulcer-related DEG analysis showed that compared to patients without ulcers, 238 genes were upregulated and 207 genes were downregulated in the ulcer samples (Figure 2). The healing-related analysis indicated that compared to the unhealed patients, 74 genes were upregulated and 28 genes were downregulated in the healed samples (Figure 3).
Functional enrichment analysis results of the ulcer-related DEGs
The results of the GO functional enrichment analysis of the ulcer-related DEGs indicated that the top 5 biological processes (BPs) of DFU were translational initiation, viral gene expression, the nuclear-transcribed messenger ribonucleic acid (mRNA) catabolic process, viral transcription, and the signal-recognition particle (SRP)-dependent co-translational protein targeting of the membrane, while the top 5 cell components (CCs) were focal adhesion, the cell-substrate junction, the cytosolic ribosome, the ribosome, and the ribosomal subunit, and the top 5 molecular functions (MFs) were the structural constituent of the ribosome, sulfur compound binding, glycosaminoglycan binding, heparin binding, and ribosomal RNA binding (Figure 4A,4B).
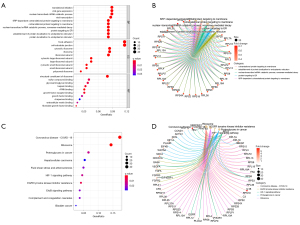
The results of the KEGG functional enrichment analysis of the ulcer-related DEGs revealed that top the top 10 enriched KEGG pathways were coronavirus disease 2019, ribosome, proteoglycans in cancer, hepatocellular carcinoma, fluid shear stress and atherosclerosis, the hypoxia inducible factor 1 (HIF-1) signaling pathway, epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor resistance, the ErbB signaling pathway, complement and coagulation cascades, and bladder cancer (Figure 4C,4D).
Functional enrichment analysis results of the healing-related DEGs
The results of the GO functional enrichment analysis of the healing-related DEGs indicated that the top 5 BPs in DFU were neutrophil degranulation, neutrophil activation involved in the immune response, leukocyte chemotaxis, cell chemotaxis, and myeloid leukocyte migration, while the top 5 CCs were the secretory granule membrane, secretory granule lumen, cytoplasmic vesicle lumen, vesicle lumen, and external side of plasma membrane, and the top 5 MFs were immune receptor activity, amide binding, peptide binding, G protein-coupled peptide receptor activity, and chemokine activity (Figure 5A,5B).
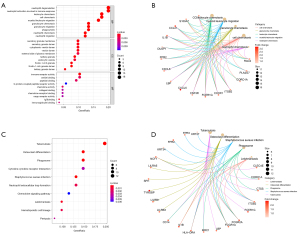
The results of the KEGG functional enrichment analysis of the healing-related DEGs revealed that the top 10 enriched KEGG pathways were tuberculosis, osteoclast differentiation, phagosome, cytokine-cytokine receptor interaction, staphylococcus aureus infection, neutrophil extracellular trap formation, chemokine signaling pathway, leishmaniasis, and hematopoietic cell lineage and pertussis (Figure 5C,5D).
Acquisition and expression analysis of DFU hub immune-related genes
We obtained the hub gene of IL-1B from the intersection genes of ulcer-related DEGs, healing-related DEGs, and immune-related genes (Figure 6A). The analysis showed that IL-1B was more highly expressed in healed DFU patients than non-healed DFU patients (Figure 6B). Additionally, IL-1B was more highly expressed in DF patients with ulcers than DF patients without ulcers (Figure 6C). These results suggest that IL-1B may play an important role in the development and healing of ulcers in DF patients.

Immune infiltration levels in DF patients
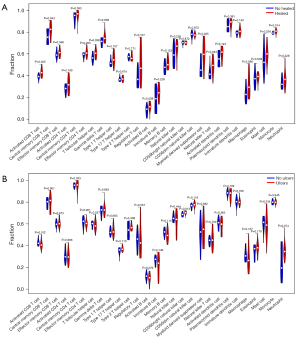
The immune infiltration analysis showed that activated cluster of differentiation (CD)8 T cells, central memory CD8 T cells, T follicular helper cells, myeloid-derived suppressor cells, natural killer T cells, and monocytes were more highly infiltrated in healed DFU patients than unhealed DFU patients (Figure 7A). CD56 bright natural killer cells were more highly infiltrated in DF patients with ulcers than DF patients without ulcers (Figure 7B).
The correlation between the expression levels of the hub gene of IL-1B and the abundance of immune cells
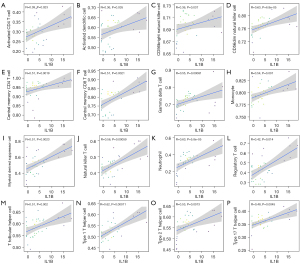
The correlation analysis showed that the expression levels of IL-1B were positively correlated with the infiltration levels of activated CD4 T cells, activated dendritic cells, CD56 bright natural killer cells, CD56 dim natural killer cells, central memory CD4 T cells, central memory CD8 T cells, gamma delta T cells, monocytes, myeloid-derived suppressor cells, natural killer T cells, neutrophils, regulatory T cells, T follicular helper cells, type 1 T helper cells, type 2 T helper cells, and type 17 T helper cells (Figure 8).
Discussion
Many diabetic patients suffer from end-point complications, such as cardiovascular disease, nephropathy, arty disease, and DF, and these complications often combine. Most DF patients suffer from ulcers in the lower limbs, and other complications can inhibit the ulcer healing process. Several studies have shown that there are various gene expression level alterations in DF and the DFU healing process (10-13). However, further research needs to be conducted to examine the fundamental biological functions of the alteration genes. We conducted the present study to investigate the DEGs between DUF, normal diabetic patients, and healed DFU patients, and examined the hub immune genes expression levels and enrichment functions. If clinicians could diagnose DF early or identify novel therapy targets, the quality of life of these patients would greatly increase, and the economic burden on society would be relieved.
Our results showed that there were 238 upregulated genes and 207 downregulated genes in the DF patients with and without ulcers, and there were 74 upregulated genes and 28 downregulated genes in the healed and unhealed samples. To understand the main biological functions of the DEGs, we performed a functional enrichment analysis. The results showed that the main biological functions of the DEGs were neutrophil degranulation, leukocyte chemotaxis, myeloid leukocyte migration, phagosome, cytokine-cytokine receptor interaction, and the chemokine signaling pathway.
Neutrophils have been proven to play a critical role in DFU (14-17). Arican et al. showed that the neutrophil-to-lymphocyte ratio can be a predict the prognosis of DFU, and a high neutrophil-to-lymphocyte ratio inhibits the healing process (15). Yang et al. also found that citH3, a neutrophil extracellular trap (NET) specific marker, acts as an inhibitor in wound healing (17). Bannon et al. found that myeloid cells were contributors of chronic inflammation during ulcer healing in mice (18). Pasquier et al. showed that osteoclast cells may have significant correlation with charcot foot disease (19). The chemokine signaling pathway has also been proven to be a significant regulator of DFU (20-22). Thus, a number of gene alterations occur during the DFU and wound healing processes. According to the functional enrichment analysis, these genes act as regulators of leukocytes and inflammation.
Ulcers occur with inflammation and immune cells, such as neutrophils, macrophages and T cells. Immune cells can secrete various chemokines to promote or inhibit inflammation. Thus, we explored the correlation between DEGs and significant immune-related genes. The intersection results showed that IL-1B was more highly expressed in DFU and healed DFU patients than patients without ulcers and unhealed DFU patients. IL-1, including IL-1α or IL-1B, is a master cytokine of local and systemic inflammation, and IL-1B is thought to act in the later phase of macrophage recruitment to damaged tissue (23). IL-1 has been shown to play a key role in various diseases, including osteoarthritis, kidney disease, and periodontitis (24-26). These diseases are always accompanied by inflammation. IL-1B and IL-1β are also regulators of the immune response (27). Only a few studies have investigated the relationship between diabetic eye disease, diabetic nephropathy, and DFU (28-30). NETs can promote the inflammation of DFUs that are regulated by IL-1B, and acts as a stimulus for NLR family pyrin domain containing 3 (NLRP3) inflammasome activation in macrophages to promote IL-1B-dependent exacerbation of inflammation, but degrading NETs improves the wound healing process (29). Thus, IL-1B may affect the DFU healing process by regulating inflammation.
Based on the importance of the role of immune cells in the occurrence and progression of DFU, we examined immune cell abundance differences among normal diabetes patients, healed DFU patients, and unhealed DFU patients. We found that activated CD8 T cells, central memory CD8 T cells, T follicular helper cells, myeloid-derived suppressor cells, natural killer T cells, and monocytes were highly infiltrated in unhealed patients. However, no difference was found between patients with and without ulcers. These results indicate that immune cells may be involved in the healing process of ulcers but not in the occurrence of ulcers. Several studies have already confirmed that immune cells or immune responses play important roles in DFU and wound healing (5,31-33). Dong et al. found that increasing the abundance of mast cells inhibited the healing process of DFU (31). Regrettably, the roles of many immune cells in the healing process of DFU have not yet been closely investigated. We also investigated the correlations between IL-1B expression levels and immune cell infiltration. The results showed that IL-1B was positively correlated with the infiltration levels of activated CD4 T cells, activated dendritic cells, CD56 bright natural killer cells, CD56 dim natural killer cells, central memory CD4 T cells, central memory CD8 T cells, gamma delta T cells, monocytes, myeloid-derived suppressor cells, natural killer T cells, neutrophils, regulatory T cells, T follicular helper cells, type 1 T helper cells, type 2 T helper cells, and type 17 T helper cells. The limitation of this study is the lack of verification of the expression and function of hub genes IL-1B.
Conclusions
In the present study, we identified DEGs in DF patients with and without ulcers, and in healed and unhealed patients. We identified a number of gene alterations, and found that the inflammation response was critical in the occurrence and healing process of DFU. The immune-related gene of IL-1B may be a diagnostic tool that can be used to assess the healing process of DFU patients. IL-1B may affect the DFU healing process by regulating inflammation and immune cell infiltration.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-75/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-75/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cole JB, Florez JC. Genetics of diabetes mellitus and diabetes complications. Nat Rev Nephrol 2020;16:377-90. [Crossref] [PubMed]
- Boulton AJM, Whitehouse RW. The Diabetic Foot. In: Feingold KR, Anawalt B, Boyce A et al., editors. Endotext. South Dartmouth (MA): MDText.com, Inc., 2000.
- Pecoraro RE, Reiber GE, Burgess EM. Pathways to diabetic limb amputation. Basis for prevention. Diabetes Care 1990;13:513-21. [Crossref] [PubMed]
- Arya AK, Tripathi K, Das P. Promising role of ANGPTL4 gene in diabetic wound healing. Int J Low Extrem Wounds 2014;13:58-63. [Crossref] [PubMed]
- Sawaya AP, Stone RC, Brooks SR, et al. Deregulated immune cell recruitment orchestrated by FOXM1 impairs human diabetic wound healing. Nat Commun 2020;11:4678. [Crossref] [PubMed]
- Wu M, Leng W, Pan H, et al. The Reduced Expression of EOLA1 May Be Related to Refractory Diabetic Foot Ulcer. Mediators Inflamm 2019;2019:6705424. [Crossref] [PubMed]
- Pradhan Nabzdyk L, Kuchibhotla S, Guthrie P, et al. Expression of neuropeptides and cytokines in a rabbit model of diabetic neuroischemic wound healing. J Vasc Surg 2013;58:766-75.e12. [Crossref] [PubMed]
- Tellechea A, Kafanas A, Leal EC, et al. Increased skin inflammation and blood vessel density in human and experimental diabetes. Int J Low Extrem Wounds 2013;12:4-11. [Crossref] [PubMed]
- Dinh T, Tecilazich F, Kafanas A, et al. Mechanisms involved in the development and healing of diabetic foot ulceration. Diabetes 2012;61:2937-47. [Crossref] [PubMed]
- Donate-Correa J, Martín-Núñez E, Ferri C, et al. FGF23 and Klotho Levels are Independently Associated with Diabetic Foot Syndrome in Type 2 Diabetes Mellitus. J Clin Med 2019;8:448. [Crossref] [PubMed]
- Januszyk M, Chen K, Henn D, et al. Characterization of Diabetic and Non-Diabetic Foot Ulcers Using Single-Cell RNA-Sequencing. Micromachines (Basel) 2020;11:815. [Crossref] [PubMed]
- Nowak WN, Borys S, Kusińska K, et al. Number of circulating pro-angiogenic cells, growth factor and anti-oxidative gene profiles might be altered in type 2 diabetes with and without diabetic foot syndrome. J Diabetes Investig 2014;5:99-107. [Crossref] [PubMed]
- Vaseghi H, Pornour M, Djavid GE, et al. Association of the gene expression variation of tumor necrosis factor-α and expressions changes of dopamine receptor genes in progression of diabetic severe foot ulcers. Iran J Basic Med Sci 2017;20:1213-9. [PubMed]
- Altay FA, Kuzi S, Altay M, et al. Predicting diabetic foot ulcer infection using the neutrophil-to-lymphocyte ratio: a prospective study. J Wound Care 2019;28:601-7. [Crossref] [PubMed]
- Arıcan G, Kahraman HÇ, Özmeriç A, et al. Monitoring the Prognosis of Diabetic Foot Ulcers: Predictive Value of Neutrophil-to-Lymphocyte Ratio and Red Blood Cell Distribution Width. Int J Low Extrem Wounds 2020;19:369-76. [Crossref] [PubMed]
- Huang W, Jiao J, Liu J, et al. MFG-E8 accelerates wound healing in diabetes by regulating "NLRP3 inflammasome-neutrophil extracellular traps" axis. Cell Death Discov 2020;6:84. [Crossref] [PubMed]
- Yang S, Gu Z, Lu C, et al. Neutrophil Extracellular Traps Are Markers of Wound Healing Impairment in Patients with Diabetic Foot Ulcers Treated in a Multidisciplinary Setting. Adv Wound Care (New Rochelle) 2020;9:16-27. [Crossref] [PubMed]
- Bannon P, Wood S, Restivo T, et al. Diabetes induces stable intrinsic changes to myeloid cells that contribute to chronic inflammation during wound healing in mice. Dis Model Mech 2013;6:1434-47. [Crossref] [PubMed]
- Pasquier J, Thomas B, Hoarau-Véchot J, et al. Circulating microparticles in acute diabetic Charcot foot exhibit a high content of inflammatory cytokines, and support monocyte-to-osteoclast cell induction. Sci Rep 2017;7:16450. [Crossref] [PubMed]
- Galstyan KO, Nedosugova LV, Martirosian NS, et al. Modification of Tumor Necrosis Factor-α and C-C Motif Chemokine Ligand 18 Secretion by Monocytes Derived from Patients with Diabetic Foot Syndrome. Biology (Basel) 2019;9:3. [Crossref] [PubMed]
- Sun X, Wang X, Zhao Z, et al. Paeoniflorin inhibited nod-like receptor protein-3 inflammasome and NF-κB-mediated inflammatory reactions in diabetic foot ulcer by inhibiting the chemokine receptor CXCR2. Drug Dev Res 2021;82:404-11. [Crossref] [PubMed]
- Wang X, Li J, Wang Z, et al. Wound exudate CXCL6: a potential biomarker for wound healing of diabetic foot ulcers. Biomark Med 2019;13:167-74. [Crossref] [PubMed]
- Afonina IS, Müller C, Martin SJ, et al. Proteolytic Processing of Interleukin-1 Family Cytokines: Variations on a Common Theme. Immunity 2015;42:991-1004. [Crossref] [PubMed]
- Anders HJ. Of Inflammasomes and Alarmins: IL-1β and IL-1α in Kidney Disease. J Am Soc Nephrol 2016;27:2564-75. [Crossref] [PubMed]
- Cheng R, Wu Z, Li M, et al. Interleukin-1β is a potential therapeutic target for periodontitis: a narrative review. Int J Oral Sci 2020;12:2. [Crossref] [PubMed]
- Jenei-Lanzl Z, Meurer A, Zaucke F. Interleukin-1β signaling in osteoarthritis - chondrocytes in focus. Cell Signal 2019;53:212-23. [Crossref] [PubMed]
- Kimura T, Jain A, Choi SW, et al. TRIM-directed selective autophagy regulates immune activation. Autophagy 2017;13:989-90. [Crossref] [PubMed]
- Buraczynska M, Ksiazek K, Wacinski P, et al. Interleukin-1β Gene (IL1B) Polymorphism and Risk of Developing Diabetic Nephropathy. Immunol Invest 2019;48:577-84. [Crossref] [PubMed]
- Lee MKS, Sreejit G, Nagareddy PR, et al. Attack of the NETs! NETosis primes IL-1β-mediated inflammation in diabetic foot ulcers. Clin Sci (Lond) 2020;134:1399-401. [Crossref] [PubMed]
- Mesquida M, Drawnel F, Fauser S. The role of inflammation in diabetic eye disease. Semin Immunopathol 2019;41:427-45. [Crossref] [PubMed]
- Dong J, Chen L, Zhang Y, et al. Mast Cells in Diabetes and Diabetic Wound Healing. Adv Ther 2020;37:4519-37. [Crossref] [PubMed]
- Ejiugwo M, Rochev Y, Gethin G, et al. Toward Developing Immunocompetent Diabetic Foot Ulcer-on-a-Chip Models for Drug Testing. Tissue Eng Part C Methods 2021;27:77-88. [Crossref] [PubMed]
- Moura J, Madureira P, Leal EC, et al. Immune aging in diabetes and its implications in wound healing. Clin Immunol 2019;200:43-54. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


