
LncRNA SNHG12 promotes the malignant progression of melanoma by targeting miR-199b
Introduction
Melanoma is a malignant tumor caused by the malignant transformation of melanocytes. It is very aggressive and has a high mortality rate (1). Recently, the incidence of melanoma has steadily increased worldwide (2). Although the current combination of surgery, chemotherapy, targeted therapy, and immunotherapy has made great progress in the treatment of melanoma, the therapeutic effect for some melanomas is still not ideal, especially for patients with distant metastases (3). The abnormal regulation of a variety of genes and signaling pathways has been associated with the occurrence of melanoma (4). Hence, it is urgent to understand the molecular mechanisms of malignant processes in melanoma and design new targets for the treatment of melanoma.
Long non-coding RNAs (lncRNAs) are non-coding RNAs with a length of at least 200 nucleotides (5). Although they do not have the ability to encode proteins, they perform a variety of biological functions (6). LncRNAs participate in a variety of carcinogenic processes such as tumor proliferation, metastasis, apoptosis inhibition, and drug resistance (7). More and more studies have shown that small nucleolar RNA host gene 12 (SNHG12) plays an important role in a variety of cancers. For instance, in diffuse large B-cell lymphoma, a high level of SNHG12 is closely associated with the poor prognosis of patients, and can accelerate tumor progression by regulating miR-195 (8). In pancreatic cancer, SNHG12 is highly expressed in cancer cell lines and tissues, and promotes the proliferation, invasion, and epithelial-mesenchymal transition of pancreatic cancer cells through regulating miR-320b (9). In breast cancer, the expression of SNHG12 is up-regulated, which can up-regulate the expression of SALL4 by inhibiting miR-15a-5p, and promotes breast cancer cell proliferation, migration, and invasion, and inhibits cell apoptosis (10). However, the role of SNHG12 in melanoma and its mechanisms remain unclear.
In the present research, we detected the expression levels of SNHG12 in melanoma cell lines. In addition, this study explored the influence of SNHG12 on the proliferation, migration, and invasion of melanoma cells in vitro. Through further research, we explained the molecular mechanisms by which SNHG12 exerts its corresponding biological functions. This study is the first to explore the role and the corresponding mechanisms of SNHG12 in melanoma, which will provide necessary theoretical basis for designing therapeutic target. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-214/rc).
Methods
Cell culture and transfection
Human melanoma cell lines (A2058, SK-MEL-28, CHL-1, and A375) and the human epidermal melanocyte cell line HEMa-LP were all from ATCC (ATCC, USA). Human melanoma cell lines were cultured in DMEM medium (Gibco, USA) containing 10% fetal bovine serum (FBS) (Invitrogen, USA). HEMa-LP cells were maintained in 254 medium (Cascade Biologics, USA). All cell lines were maintained in a 37 °C cell incubator containing 5% CO2. Cell transfection was performed with Lipofectamine 3000 (Invitrogen).
Quantitative real-time PCR (qRT-PCR) experiment
TRIzol reagent (Invitrogen) was used to isolate RNA from cells. Subsequently, ribonucleic acid (RNA) was reverse transcribed into cDNA using a cDNA synthesis kit (Takara Biotechnology, Japan). The PCR reaction was then conducted on the ABI 7500 rapid PCR system (Applied Biosystems, USA) using a SYBR Green Master Mix (Takara Biotechnology). The internal references of SNHG12 and miR-199b were GAPDH and U6, respectively. All reactions were repeated at least 3 times. Finally, the 2−ΔΔCq method was applied to analyze the results, and the relative levels of SNHG12 and miR-199b were calculated.
CCK-8 experiment
Cell proliferation ability was measured with a CCK-8 kit (Beyotime, China). A total of 5×103 cells were seeded in a 96-well plate and cultured for 24 hours before transfection. After transfecting for 24, 48, and 72 hours, 10 µL CCK-8 solution was added into each well and incubated for an appropriate time. Subsequently, a spectrophotometer was used to detect the absorbance at 450 nm.
EdU experiment
Cells were seeded and cultured for 24 hours before transfection. An EdU imaging kit (Life Technologies, USA) was used to detect DNA synthesis in melanoma cells. Cell immunostaining was observed with a fluorescence microscope, and the cells positive for EdU staining were counted.
Scratch test
Cells were seeded and cultured for 24 hours before transfection. When the cells reached approximately 90% confluence, the cell monolayer was scratched with a 10 µl pipette tip to form a uniform scratch, and the cells were rinsed twice with serum-free DMEM. The cells were then cultured in serum-free DMEM for 24 hours. Subsequently, an optical microscope (Olympus, Japan) was used to take pictures of the scratches, the fusion of the scratches was recorded, and the cell migration rate was calculated.
Transwell experiment
The Transwell test was performed using an 8.0 µm Transwell plate (Corning, USA) covered with a matrix gel. The cells were resuspended in 200 µL of serum-free DMEM medium and put into the upper chamber of the Transwell, and 500 µL of DMEM medium containing 10% FBS was put into the lower chamber. The cells above the membrane were wiped off with a cotton swab after culturing for 24 hours, and cells under the membrane were fixed with methanol at room temperature and stained with 0.1% crystal violet solution. The invading cells in 5 random fields were then observed with an optical microscope and counted.
Luciferase reporter gene experiment
The SNHG12 and ETS1 3'UTR fragments containing miR-199b binding sites were constructed into the pMIR-REPORT reporter gene vector, and the mutant plasmid served as a control. The miR-199b mimics were co-transfected into melanoma cells together with luciferase reporter gene plasmids. The luciferase activity was detected with the dual luciferase reporter gene detection system (Promega, USA).
Western blotting
After the cells were transfected, the cells were lysed with RIPA buffer (Cell Signaling Technology, USA), and the protein concentration was measured using the BCA protein quantification kit (Thermo Fisher Scientific, USA). The same amount of protein (20 µg) was separated by SDS-PAGE electrophoresis and then transferred to the polyvinylidene fluoride (PVDF) membrane. After blocking with 5% skimmed milk at room temperature for 1 hour, the membrane was incubated with the primary antibody at 4 °C overnight, and then with the secondary antibody at room temperature for 2 hours. Finally, the ECL Luminescence Kit (Invitrogen) and gel imaging system (Bio-Rad Laboratories, USA) were used for imaging. The antibodies used in this experiment were anti-ETS1 antibody (ab26096, Abcam, USA), anti-Paxillin antibody (ab32084, Abcam), anti-Jagged1 antibody (ab7771, Abcam), and anti-DDR1 antibody (#5583, Cell Signaling Technology). GAPDH was used as the protein internal control.
Statistical analysis
SPSS 13.0 was used to analyze the data, and all experiments were repeated independently no less than 3 times. All data were presented as mean ± standard deviation. The Student’s t-test was performed to analyze the differences between the 2 groups, and comparisons between 3 or more groups were carried out by one-way analysis of variance. P value <0.05 indicated that the difference was statistically significant.
Results
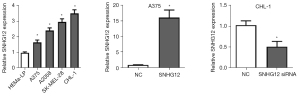
SNHG12 is highly expressed in melanoma cell lines
First, qRT-PCR was used to detect the expression levels of SHNG12 in melanoma cell lines. Compared with HEMa-LP cells, melanoma cell lines also showed higher SNHG12 expression levels (Figure 1A). Since the expression of SNHG12 in A375 cells was lower while SNHG12 expression in CHL-1 cells was higher, an SNHG12 expression plasmid was transfected in A375 cells to overexpress SNHG12, and siRNA was transfected in CHL-1 cells to silence SNHG12 (Figure 1B,1C).
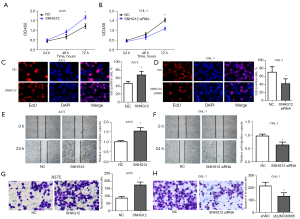
SNHG12 promotes the proliferation, migration, and invasion of melanoma cells
In order to study the biological functions of SNHG12, after overexpressing SNHG12 in A375 cells and silencing SNHG12 in CHL-1 cells, CCK-8 and EdU experiments were performed to study the influence of SNHG12 on cell proliferation (Figure 2A-2D), the scratch assays were applied to study the influence of SNHG12 on cell migration (Figure 2E,2F), and the Transwell experiment was conducted to study the influence of SNHG12 on cell invasion (Figure 2G,2H). The results demonstrated that after overexpressing SNHG12 in A375 cells, cell proliferation was significantly accelerated (Figure 2A,2C), and the migration and invasion capabilities were significantly enhanced (Figure 2E,2G). Correspondingly, after silencing SNHG12 in CHL-1 cells, the proliferation of the cells was significantly slowed down (Figures 2B,2D), and the migration and invasion capabilities were significantly weakened (Figures 2F,2H). These results demonstrated that SNHG12 was able to promote the proliferation, migration, and invasion of melanoma cells in vitro.
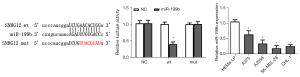
MiR-199b is the target gene of SNHG12
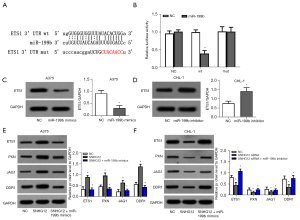
Through the starbase database, we speculated that miR-199b might be the target gene of SNHG12 (Figure 3A). According to this predicted site, pMIR-SNHG12-wt and pMIR-SNHG12-mut luciferase reporter gene plasmids were constructed respectively, and co-transfected with NC or miR-199b mimics into melanoma cells. The results showed that when miR-199b mimics were co-transfected with pMIR-SNHG12-wt, the relative activity of luciferase was significantly reduced (Figure 3B), which indicated that miR-199b was the target gene of SNHG12. Moreover, our results showed that miR-199b was under-expressed in melanoma cell lines (Figure 3C).
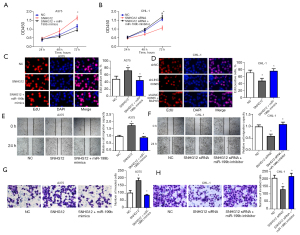
SNHG12 regulates the proliferation, migration, and invasion of melanoma cells through miR-199b
CCK-8 assays, EdU assays, scratch assays and Transwell assays were performed to study the influence of SNHG12 and miR-199b on cell proliferation, migration and invasion (Figure 4A-4H). Transfection of miR-199b mimics in A375 cells overexpressing SNHG12 attenuated the promotive effect of SNHG12 on cell proliferation, migration, and invasion (Figure 4A,4C,4E,4G). Transfection of an miR-199b inhibitor in SNHG12-silenced CHL-1 cells restored the reduced cell proliferation, migration, and invasion ability caused by SNHG12 knockdown (Figure 4B,4D,4F,4H). The above results suggested that SNHG12 regulates the proliferation, migration, and invasion of melanoma cells through miR-199b.
SNHG12 regulates the expression of miR-199b target genes
Through the starbase database, it was found that the 3'UTR of ETS1 has a binding site for miR-199b (Figure 5A). According to this predicted site, pMIR-ETS1 3'UTR-wt and pMIR-ETS1 3'UTR-mut luciferase reporter gene plasmids were constructed respectively, and co-transfected with NC or miR-199b mimics into melanoma cells. The results showed that the activity of the wild-type luciferase plasmid was inhibited by miR-199b mimics (Figure 5B). At the same time, overexpression of miR-199b could inhibit the expression of ETS1 at the protein level, while silencing miR-199b promoted the expression of ETS1 protein (Figure 5C,5D). The above results suggested that ETS1 was the target gene of miR-199b, and SNHG12 regulated the expression of ETS1 through miR-199b (Figure 5E,5F). Moreover, SNHG12 could regulate the expression of other proven target genes of miR-199b such as PXN, JAG1, and DDR1 (Figure 5E,5F).
Discussion
Recent studies have shown that lncRNA-miRNA regulatory networks are widely implicated in the pathogenesis of tumors (11). LncRNAs become key regulators of malignant tumors by combining with miRNAs to regulate gene expression, which is a common process of regulation (12). More and more studies have proven that lncRNAs and miRNAs play vital roles in the occurrence and development of melanoma, and may be useful biomarkers and therapeutic targets in the future (13). For example, the expression of lncRNA FGD5-AS1 in melanoma tissue was significantly up-regulated, and its high expression was significantly related to tumor thickness, tumor stage, and overall survival rate, and may be a potential independent prognostic factor for melanoma (14). LncRNA FOXC2-AS1 is up-regulated in melanoma tissues, especially in patients with metastasis or stage II-IV melanoma, and patients with high FOXC2-AS1 expression levels have lower survival rates. FOXC2-AS1 can stimulate the proliferation of melanoma by silencing p15 through recruiting EZH2 (15). Moreover, lncRNA SNHG16 is highly expressed in melanoma and facilitates melanoma cell proliferation and migration by regulating miR-132 (16).
LncRNA SNHG12 is located on chromosome 1p35.3 and is related to a variety of cancers. However, its biological role and related mechanisms in melanoma are still unclear. In this study, it was confirmed that the expression of SNHG12 in melanoma cell lines was obviously up-regulated, suggesting that SNHG12 may play vital roles in the progression of melanoma. Subsequently, the biological functions of SNHG12 in melanoma were evaluated, and the results showed that overexpressing the SNHG12 gene obviously facilitated the proliferation, migration, and invasion of melanoma cells. In contrast, silencing SNHG12 impeded the proliferation, migration, and invasion of melanoma cells. Therefore, it is believed that SNHG12 plays an oncogenic role in melanoma.
Bioinformatics analysis was then used to find a binding site of miR-199b in SNHG12, and further study the regulatory mechanism of SNHG12 on miR-195. It was confirmed for the first time that miR-199b is a target gene of SNHG12, and SNHG12 can affect the proliferation, migration, and invasion of melanoma cells via regulating miR-199b. According to reports, miR-199b is related to a variety of cancers, and can inhibit cancer progression by directly targeting and regulating certain oncogenes such as PXN, JAG1, and DDR1 (17-19). In melanoma, miR-199b is very effective in inhibiting the proliferation and viability of melanoma cells, affecting key signaling pathways related to melanoma cell survival, and enhancing the efficacy of drugs to inhibit BRAF and MEK (20). Furthermore, this study found that ETS1 was a new target gene of miR-199b, and miR-199b was able to directly target and reduce the expression of ETS1 in melanoma. ETS1 is a member of the ETS transcription factor family, and acts as a transcriptional activator and suppressor of many genes, participating in the development of stem cells, cell senescence, and death, as well as tumorigenesis (21). In most cancers, the expression of ETS1 is related to low survival rates. ETS1 helps to enhance the invasiveness of cancer cells, promote the transformation of epithelial cells to mesenchymal cells, and promote drug resistance and angiogenesis (22). This study further validated that SNHG12 can promote the expression of miR-199b target genes (ETS1, PXN, JAG1, and DDR1) by regulating miR-199b.
This study found that SNHG12 can promote the malignant progression of melanoma as an oncogene. Therefore, drugs targeting SNHG12 may have potential application value for the clinical treatment and prognosis of melanoma. However, there are still many deficiencies in this study. For example, this study did not involve in vivo data and signaling pathways, which might be very promising in this field.
Conclusions
LncRNA SNHG12 is overexpressed in melanoma cell lines, and can promote the expression of miR-199b target genes ETS1, PXN, JAG1, and DDR1 through targeted regulation of miR-199b, thereby promoting the proliferation, migration, and invasion of melanoma cells.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-214/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-214/dss
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-214/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rebecca VW, Somasundaram R, Herlyn M. Pre-clinical modeling of cutaneous melanoma. Nat Commun 2020;11:2858. [Crossref] [PubMed]
- Schadendorf D, van Akkooi ACJ, Berking C, et al. Melanoma. Lancet 2018;392:971-84. [Crossref] [PubMed]
- Grob JJ, Garbe C, Ascierto P, et al. Adjuvant melanoma therapy with new drugs: should physicians continue to focus on metastatic disease or use it earlier in primary melanoma? Lancet Oncol 2018;19:e720-e725. [Crossref] [PubMed]
- Bai X, Fisher DE, Flaherty KT. Cell-state dynamics and therapeutic resistance in melanoma from the perspective of MITF and IFNγ pathways. Nat Rev Clin Oncol 2019;16:549-62. [Crossref] [PubMed]
- Nair L, Chung H, Basu U. Regulation of long non-coding RNAs and genome dynamics by the RNA surveillance machinery. Nat Rev Mol Cell Biol 2020;21:123-36. [Crossref] [PubMed]
- Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol 2019;21:542-51. [Crossref] [PubMed]
- Tan H, Zhang S, Zhang J, et al. Long non-coding RNAs in gastric cancer: New emerging biological functions and therapeutic implications. Theranostics 2020;10:8880-902. [Crossref] [PubMed]
- Chen LY, Zhang XM, Han BQ, et al. Long Noncoding RNA SNHG12 Indicates the Prognosis and Accelerates Tumorigenesis of Diffuse Large B-Cell Lymphoma Through Sponging microR-195. Onco Targets Ther 2020;13:5563-74. [Crossref] [PubMed]
- Cao W, Zhou G. LncRNA SNHG12 contributes proliferation, invasion and epithelial-mesenchymal transition of pancreatic cancer cells by absorbing miRNA-320b. Biosci Rep 2020;40:BSR20200805. [Crossref] [PubMed]
- Yuan JH, Li WX, Hu C, et al. Upregulation of SNHG12 accelerates cell proliferation, migration, invasion and restrain cell apoptosis in breast cancer by enhancing regulating SALL4 expression via sponging miR-15a-5p. Neoplasma 2020;67:861-70. [Crossref] [PubMed]
- Fu S, Wang Y, Li H, et al. Regulatory Networks of LncRNA MALAT-1 in Cancer. Cancer Manag Res 2020;12:10181-98. [Crossref] [PubMed]
- Yang Y, Wang Y, Wang F, et al. The roles of miRNA, lncRNA and circRNA in the development of osteoporosis. Biol Res 2020;53:40. [Crossref] [PubMed]
- Safa A, Gholipour M, Dinger ME, et al. The critical roles of lncRNAs in the pathogenesis of melanoma. Exp Mol Pathol 2020;117:104558. [Crossref] [PubMed]
- Gao Y, Zhu H, Mao Q. Expression of lncRNA FGD5-AS1 correlates with poor prognosis in melanoma patients. J Gene Med 2020;22:e3278. [Crossref] [PubMed]
- Xu DF, Tao XH, Yu Y, et al. LncRNA FOXC2-AS1 stimulates proliferation of melanoma via silencing p15 by recruiting EZH2. Eur Rev Med Pharmacol Sci 2020;24:8940-6. [PubMed]
- Bi LL, Hua XQ, Li WH, et al. SNHG16 promotes cell proliferation and migration through sponging miR-132 in melanoma. J Biol Regul Homeost Agents 2020;34:1307-16. [PubMed]
- Du C, Wang Y, Zhang Y, et al. LncRNA DLX6-AS1 Contributes to Epithelial-Mesenchymal Transition and Cisplatin Resistance in Triple-negative Breast Cancer via Modulating MiR-199b-5p/Paxillin Axis. Cell Transplant 2020;29:963689720929983. [Crossref] [PubMed]
- Chen LY, Zhi Z, Wang L, et al. NSD2 circular RNA promotes metastasis of colorectal cancer by targeting miR-199b-5p-mediated DDR1 and JAG1 signalling. J Pathol 2019;248:103-15. [Crossref] [PubMed]
- Wu A, Chen Y, Liu Y, et al. miR-199b-5p inhibits triple negative breast cancer cell proliferation, migration and invasion by targeting DDR1. Oncol Lett 2018;16:4889-96. [Crossref] [PubMed]
- Fattore L, Campani V, Ruggiero CF, et al. In Vitro Biophysical and Biological Characterization of Lipid Nanoparticles Co-Encapsulating Oncosuppressors miR-199b-5p and miR-204-5p as Potentiators of Target Therapy in Metastatic Melanoma. Int J Mol Sci 2020;21:1930. [Crossref] [PubMed]
- Shaikhibrahim Z, Wernert N. ETS transcription factors and prostate cancer: the role of the family prototype ETS-1. Int J Oncol 2012;40:1748-54. [PubMed]
- Dittmer J. The role of the transcription factor ETS1 in carcinoma. Semin Cancer Biol 2015;35:20-38. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


