Construction of ceRNA networks with different types of IDH1 mutation status in low-grade glioma patients
Introduction
Glioma is the most common brain carcinoma, and is divided into low-grade glioma (LGG) and high-grade glioma (HGG) (1). Under the classification system of the World Health Organization (WHO), LGG includes grade II and III gliomas (2). The 5-year survival rate of LGG is significantly higher than that of HGG, which can reach 80% (3).
In recent years, molecular diagnosis has become an integral part of LGG diagnosis. About 75% of LGG patients carry isocitrate dehydrogenase 1 (IDH1) mutation and that IDH1 mutation status is associated with the prognosis of LGG patients (4,5), so a complete diagnosis and prognostic assessment of gliomas should include IDH1 mutation status (6,7). IDH1 is a key enzyme in the glucose metabolism pathway, which can catalyze the oxidative decarboxylation of isocitrate to α-ketoglutarate, and protect cells from oxidative stress (4). It was proved that high glucose metabolism status promotes glioma cell growth by upregulating the expression and function of growth factor receptors (8). Berghoff et al. (9) found that IDH1 mutation status in LGG is closely related to the tumor immune microenvironment. Patients with mutated IDH1 have higher immune cell infiltration and higher programmed cell death-ligand 1 (PD-L1) expression levels than patients with non-mutated IDH1 (9). A previous study has found evidence of an association between IDH1 mutation status and cluster of differentiation (CD)8+ T cells or immune responses (10).
Salmena et al. (11) proposed the competitive endogenous RNA (ceRNA) hypothesis, which holds that different ribonucleic acids (RNAs), such as messenger RNA (mRNA), long non-coding RNA (lncRNA), and other pseudogenes competitively combine with the corresponding micro RNA (miRNA) to form a large-scale regulatory network. This endogenous competitive relationship can affect the biological behavior of tumors. LncRNAs are RNAs >200 bp in length that do not encode proteins (12,13). LncRNAs serve as ceRNAs, share a common functionality in regulating gene expression and encoding miRNAs, and have important roles in oncogenesis and tumor progression (12,13).
In this paper, we aimed to construct a ceRNA network to identify differentially expressed genes (DEGs) between patients’ samples containing mutated IDH1 and wild-type IDH1 gene variants. We believe that our study makes a significant contribution to the literature because we conducted the first ever large-scale analysis of differential gene expression in glioma patients with differential genetic background establishing ceRNA regulatory networks. We hope that this information will be helpful in identifying immune cell infiltration-associated mRNAs and lncRNAs as potential treatment targets for patients with LGG containing wild-type IDH1 gene variants.
We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6983/rc).
Methods
Patient data
A total of 30 pairs of LGG tissue samples and adjacent normal tissue samples were obtained from patients at the Affiliated Suzhou Hospital of Nanjing Medical University; none of the patients had undergone radiotherapy or chemotherapy before surgery. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of the Affiliated Suzhou Hospital of Nanjing Medical University (No. KL901199) and informed consent was taken from all the patients.
Data collection and analysis
RNA sequencing (RNA-Seq) data and the corresponding clinical information of the LGG patients was obtained from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/repository) and Chinese Glioma Genome Atlas (CGGA) data portal (http://www.cgga.org.cn/download.jsp). The mRNA, lncRNA, and miRNA sequence information was obtained from the Illumina HiSeqmiRNASeq and Illumina HiSeqRNASeq platforms. Five hundred and thirty LGG tissue samples and 5 normal samples were obtained from TCGA, and 139 LGG tissue samples and 4 normal samples were obtained from CGGA. We obtained a list of immune-related genes from the Immunology Database and Analysis Portal system (ImmPort, https://www.immport.org).
Identification of DERNA
The method used to identify DEGs was the same as that described in our previous article (14). The DEGs were allocated to the following two groups: (I) the different IDH1 mutation status group (lncRNA (DEIDH1lncRNA), mRNA (DEIDH1mRNA), and miRNA (DEIDH1miRNA)); and (II) the LGG normal group (lncRNA (DELGGlncRNA), and mRNA (DELGGmRNA)). The cutoff criteria were as follows; |log2 fold Change| >1.0, and a P value <0.05.
Establishment of the ceRNA network
The relationship between lncRNA and miRNA was identified by the miRcode database, and the sequence information of the miRNAs was obtained from the StarBase v2.0 database. The TargetScan and miRDB databases provided the investigators with information about the miRNA and mRNA interactions (15,16). mRNAs that could be searched in both databases were defined as the candidate mRNAs. Next, the candidate mRNAs were intersected with the differentially expressed mRNAs to identify the mRNAs targeted by the DEmiRNAs. Next, a ceRNA network of DEGs based on the DEmiRNA-DElncRNA and DEmiRNA-DEmRNA interactions was established using Cytoscape 3.7.2 (17,18).
Functional enrichment analysis
The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes analyses were conducted using R package to clarify the functional enrichment of genes in the ceRNA network. The cutoff P value for the GO and KEGG analyses was <0.05.
Survival analysis
We identified the prognostic mRNA, LncRNA, and miRNA based on a univariate Cox analysis. For a further analysis of the survival prognosis, we constructed Kaplan-Meier plots, and used the log-rank test for the statistical analysis.
The immune cell infiltration status of TCGA LGG patients
We downloaded the immune cell infiltration data of LGG patients from the TIMER website (https://cistrome.shinyapps.io/). TIMER is an open website that contains over 10,000 samples of about 30 cancer types with 6 types of immune cell infiltration from TCGA, including CD8+ T cells, B cells, CD4+ T cells, neutrophils, macrophages, and dendritic cells (DCs) (19).
Quantitative real-time PCR
Total RNA was isolated from the samples of 30 LGG patients by TRIzol reagent (Invitrogen, USA), and reverse transcribed using a First Strand cDNA Synthesis Kit (New England Biolabs, China). RNA amplification was performed using a SYBR Green polymerase chain reaction (PCR) kit (Applied Biological Materials, Canada) based on the Applied Biosystems 7500Real-Time PCR System (Applied Biosystems, USA). The 2−ΔΔCt method was used to normalize the expression of RNA. The PCR primers in this study are shown in Table S1; three independent experiments were conducted.
Statistical analyses
All the statistical analyses were performed using SPSS 23.0 (Chicago, USA) and GraphPad Prism 8.0 (San Diego, USA) software. A Spearman’s rank analysis was conducted to examine the association between gene expression and immune cell infiltration. The Student’s t-test was used to analyze differences between the groups, and a univariate Cox regression analysis was conducted to identify the prognostic genes. A P value <0.05 was considered statistically significant.
Results
Clinical features of LGG patients
A total of 30 LGG patients were included in our study, of whom 15 had IDH1 mutations and 15 had wild-type IDH1gene variants. The clinical features of TCGA and CGGA patients are shown in Table 1. Of the 530 TCGA patients, patients with no overall survival (OS) data and those with IDH1 mutation status were removed, and subsequently, our research database comprised 506 patients (225 female and 281 male, with a median age of 41 (range, 14–87 years). Two hundred and forty-three patients had grade II LGG, and 263 had grade III LGG. Three hundred and eighty-nine patients had the IDH1 mutation, and 117 patients had the wild-type IDH1 form. The wild-type IDH1 patients had worse OS than the IDH1 mutation patients (see Figure S1A).
Table 1
| Clinical features | TCGA (N=506) | CGGA (N=182) |
|---|---|---|
| Age (years) | ||
| Median | 41 | 39 |
| Range | 14–87 | 10–74 |
| Gender, n (%) | ||
| Female | 225 (44.47) | 71 (39.01) |
| Male | 281 (55.53) | 111 (60.99) |
| Grade, n (%) | ||
| G2 | 243 (48.02) | 103 (56.59) |
| G3 | 263 (51.98) | 79 (43.41) |
| IDH1 mutation, n (%) | ||
| Mutant | 389 (76.88) | 133 (73.08) |
| Wild-type | 117 (23.22) | 48 (26.37) |
| Unknown | 0 | 1 (0.55) |
LGG, low-grade glioma; CGGA, Chinese Glioma Genome Atlas; IDH1, isocitrate dehydrogenase 1; LGG, low-grade glioma; TCGA, The Cancer Genome Atlas.
The CGGA validation data set comprised 182 patients with primary LGG. Among them, 71 were female and 111 were male. The patients had a median age of 39 (range, 10–74) years. One hundred and three patients had grade II LGG and 79 had grade III LGG. 133 patients had a mutation in the IDH1 gene, 48 had a normal form of IDH1, and 1 had unknown mutational status. The wild-type IDH1 patients had worse OS than the IDH1 mutation patients (see Figure S1B).
Identified DEIDH1lncRNAs, DEIDH1miRNAs, and DEIDH1mRNAs in LGG patients with different IDH1 mutation statuses
The DEGsIDH1 for 113 LGG tissue samples with wild-type IDH1 and the DEGs for 398 LGG tissue samples with mutated IDH1 were identified as significant by the “DESeq” R package. After the analysis, we identified 2,196 DEIDH1mRNAs (302 upregulated and 1,894 downregulated) (Figure 1A), 1,294 DEIDH1lncRNAs (328 upregulated and 966 downregulated) (Figure 1B), and 29 DEIDH1miRNAs (7 upregulated and 22 downregulated) (Figure 1C). The distribution of all DEGs in the 2 ranges of -log (false discovery rate (FDR)) and logFC are shown in the volcano map in Figures 1A-1C. The heatmap is shown in Figure 1D-1F.

Establishment of the ceRNA network
To analyze the mechanism by which lncRNA mediates mRNA regulation by binding to miRNAs in LGG patients with different IDH1 mutational statuses, a network containing related lncRNAs, miRNAs, and mRNAs (the ceRNA network) was established and visualized by Cytoscape. A total of 1,294 DEIDH1lncRNAs were obtained from the miRcode database, and 177 pairs of interacting lncRNAs and miRNAs were identified by the Perl program. The target mRNAs of the 6 miRNAs were identified by the TargetScan and miRDB databases. The mRNAs included in the two gene sets were finally selected, and the mRNAs excluded in the DEmRNA gene set were removed. As a result, 88 DEIDH1mRNAs were identified in the ceRNA network (see Figure S2). Overall, a total of 88 DElncRNAs, 6 DEmiRNAs, and 88 DEmRNAs were included in the ceRNA network (see Figure 2).

Functional enrichment analysis of DEmRNAs in the ceRNA network
The biological functions of 88 DEIDH1mRNAs were examined via GO and KEGG analyses. A total of 38 biological process categories were identified in the GO analysis (P<0.05), and 5 significantly enriched pathways were found (see Figure 3A and Table S2). The most enriched GO term was the “transcription, DNA-template”. Additionally, 8 significant pathways were identified in the KEGG pathway analysis (see Figure 3B and Table S3). The most enriched KEGG pathway was the “phosphatidylinositol 3'-kinase (PI3K)-Akt signaling pathway.”

Survival-related lncRNAs in the ceRNA network
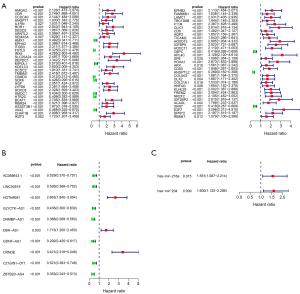
To explore the relationship between DEIDH1RNAs and the prognosis of patients with gliomas, a prognostic signature was established based on the univariate Cox regression analysis. Consequently, 65 DEIDH1mRNAs, 10 DEIDH1lncRNAs, and 2 DEIDH1miRNAs were found to be significantly related to the outcomes of LGG patients (see Figure 4A-4C).
Identification of the common DELGGlncRNAs and common immune-related DELGGmRNAs, and the establishment of the relevant ceRNA network
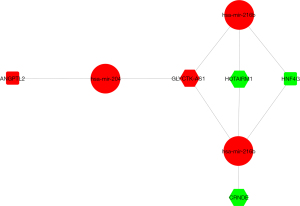
To search further for potential therapeutic targets for LGG, we identified the DELGGlncRNAs and DELGGmRNAs in the LGG tissue samples and adjacent tissue samples (see Figure S2). The heatmaps and volcano plots of DELGGmRNAs or DELGGlncRNAs in LGG and normal tissue samples were shown in Figure S3. Next, we interpolated the DEmRNAs, the prognostic-related DEIDH1mRNAs, and the immune-related genes to obtain 2 mRNAs [i.e., Hepatocyte nuclear factor 4 (HNF4G) and angiopoietin like 2 (ANGPTL2)] (see Figure S4A). These 2 genes may be related to LGG immunity. Five lncRNAs (i.e., the colorectal neoplasia differentially expressed (CRNDE), HOXA transcript antisense RNA, myeloid-specific 1 (HOTAIRM1), GLYCTK antisense RNA 1 (GLYCTK-AS1), ZBTB20 antisense RNA 4 (ZBTB20-AS4), and long intergenic non-protein coding RNA 519 (LINC00519)) were identified by intersecting the DELGGlncRNAs and the prognosis-related lncRNAs (see Figure S4B). Finally, we compared the obtained mRNAs and lncRNAs with the ceRNA network (see Figure 2). We found that the mRNA HNF4G and ANGPTL2 and the lncRNA CRNDE, HOTAIRM1, and GLYCTK-AS1 may interact via miRNA204, miRNA216ai, and miRNA216b, and thus constructed a new network (see Figure 5).
Validation of gene expression and prognosis in the ceRNA network
The expression and survival analysis of related genes were verified in the CGGA database. First, we verified gene expression in the CGGA database (see Figure 6A-6E). The expression levels of HNF4G, CRNDE, and HOTAIRM1 in patients with mutated IDH1 were lower than those of patients with wild-type IDH1, while the expression levels of ANGPTL2 and GLYCTK-AS1 in patients with mutated IDH1 were higher than those of patients with wild-type IDH1.

Second, we verified survival in the CGGA database. We divided the patients into two groups based on median gene expression. The higher expression group of HNF4G (see Figure 7A), HOTAIRM1 (see Figure 7B), and CRNDE (see Figure 7C) had worse OS than the lower expression group. The lower expression group of ANGPTL2 (see Figure 7D) had worse OS than the higher expression group. The survival analysis of these 4 genes was consistent with TCGA database. However, there was no statistically significant difference in the survival analysis of GLYCTK-AS1 (see Figure 7E). Next, we divided patients into the following four groups: (I) the low expression and IDH1 mutant group; (II) the high expression and IDH1 mutant group; (III) the low expression and IDH1 wild-type group; and (IV) the high expression and IDH1 wild-type group. The high expression and IDH1 wild-type group of HNF4G (see Figure 7F), HOTAIRM1 (see Figure 7G), CRNDE (see Figure 7H) had worse OS than the other 3 groups. The low expression and IDH1 wild-type group of ANGPTL2 (see Figure 7I) had worse OS than the other groups.

Finally, we measured expression levels using LGG patients’ tissue samples and normal tissue samples. The expression levels of HNF4G (see Figure 8A), CRNDE (see Figure 8B), and HOTAIRM1 (see Figure 8C) were consistent with those in TCGA and CGGA databases. The expression levels of ANGPTL2 (see Figure 8D) and GLYCTK-AS1 (see Figure 8E) were inconsistent with those in TCGA and CGGA databases.

Through further analysis we found that mRNA HNF4G, lncRNA CRNDE, and HOTAIRM1 were consistent in both databases with our samples. HNF4G expression was positively correlated with CRNDE expression in both TCGA (r=0.447, P<0.001) (see Figure S5A) and CGGA (r=0.482, P<0.001; see Figure S5B) databases. HNF4G expression was positively related to HOTAIRM1 expression in both TCGA (r=0.343, P<0.001; see Figure S5C) and CGGA (r=0.370, P<0.001; see Figure S5D) databases.
The relationship between different IDH1 mutation statuses and the expression of key genes with immune cell infiltration and PD-L1 expression
To further clarify the relationship between IDH1 mutation status and the immune microenvironment and PD-L1 expression, we analyzed the 6 immune cell infiltration levels in LGG patients by comparing mutated IDH1 and wild-type IDH1. The LGG patients with normal IDH1 had higher infiltration levels of 5 types of immune cells; that is, B cells (see Figure 9A), DCs (see Figure 9B), CD8+ T cells (see Figure 9C), neutrophils (see Figure 9D), and macrophages (see Figure 9E). CD4+ T cell (see Figure 9F) infiltration was not significantly different between the two groups of patients. The PD-L1 expression levels were higher in LGG patients with wild-type IDH1 than those with mutant IDH1 in TCGA (see Figure 9G) and CGGA (see Figure 9H) databases.

The relationship between expression of immune-related genes and DElncRNAs with immune cell infiltration are shown in Figure 10.

For mRNA, HNF4G expression was positively correlated with B cells (r=0.179, P<0.001; see Figure 10A), CD4+ T cells (r=0.098, P=0.028; see Figure 10B), CD8+ T cells (r=0.197, P<0.001; see Figure 10C), neutrophils (r=0.202, P=0.001; see Figure 10D), macrophages (r=0.189, P<0.001; see Figure 10E), and DCs (r=0.199, P<0.001; see Figure 10F).
For lncRNA, HOTAIRM1 expression was positively correlated with B cells (r=0.211, P<0.001; see Figure 10G), CD4+ T cells (r=0.101, P=0.022; see Figure 10H), CD8+ T cells (r=0.228, P<0.001; see Figure 10I), neutrophils (r=0.240, P=0.001; see Figure 10J), macrophages (r=0.245, P<0.001; see Figure 10K), and DCs (r=0.226, P<0.001; see Figure 10L). CRNDE expression was positively correlated with B cells (r=0.301, P<0.001; see Figure 10M), CD4+ T cells (r=0.161, P<0.001) (see Figure 10N), CD8+ T cells (r=0.263, P<0.001) (see Figure 10O), neutrophils (r=0.281, P=0.001; see Figure 10P), macrophages (r=0.247, P<0.001; see Figure 10Q), and DCs (r=0.308, P<0.001; see Figure 10R).
The expression of PD-L1 was positively related to the expression of HNF4G (r=0.332, P<0.001; see Figure 11A), CRNDE (r=0.240, P<0.001; see Figure 11B), and HOTAIRM1 (r=0.178, P<0.001; see Figure 11C) in TCGA. The expression of PD-L1 was positively related to HNF4G (r=0.285, P<0.001; see Figure 11D), however, the expression correlation between PD-L1 with CRNDE (r=0.132, P=0.075; see Figure 11E), and HOTAIRM1 (r=0.082, P=0.273; see Figure 11F) was not statistically significant in the CGGA database.

Discussion
In this article, the DEGs in LGGs with different IDH1 mutational statuses were analyzed, and the following were identified: 2,196 DEIDH1mRNAs, 1,294 DEIDH1lncRNAs, and 29 DEIDH1miRNAs. From these DEGs, we established a ceRNA network. Our ceRNA network comprised 88 DEIDH1mRNAs, 88 DEIDH1lncRNAs, and 6 DEIDH1miRNAs (see Figure 2). Through the univariate Cox regression analysis, we identified 65 mRNAs and 10 lncRNAs that correlated with prognosis in the ceRNA network (see Figure 4). The functional enrichment analysis indicated that the highest enrichment of DEIDH1mRNAs in the network was for the PI3K-Akt signaling pathway and transcription-DNA-template (see Figure 3).
Despite the rapid development of the molecular detection field in recent years, IDH1 mutation status is still one of the most stable detection markers in gliomas (20). Kloosterhof et al. (4) found that IDH1 mutation status is an important prognostic factor for gliomas. In TCGA and CGGA databases, LGG patients with mutated IDH1 have worse OS than those with the normal form of IDH1. Previously, immune cell infiltration and PD-L1 expression were thought to be related to different types of glioma (21). However, recent research has shown that IDH1 mutation status is also closely related to the immune microenvironment of gliomas. Berghoff et al. (9) showed that patients with gliomas containing wild-type IDH1 had more prominent tumor infiltrating lymphocyte infiltration and higher PD-L1 expression than patients with mutant IDH1. Amankulor et al. found IDH1 mutations down-regulated leukocyte chemotaxis level and then suppressed the tumor-related immune system (22). Additionally, IDH1 mutation in glioma mediated natural killer (NK) cell resistance by epigenetic silencing of NK group 2D (NKG2D) (23). The analysis of LGG patients in TCGA database revealed that the infiltration of multiple immune cells in patients with wild-type IDH1 was higher than that in those with mutated IDH1. Further, according to the information obtained from TCGA and CGGA databases, the expression of PD-L1 in patients with wild-type IDH1 was significantly higher than that of patients with mutated IDH1. This may indicate that different IDH1 mutation statuses may produce different immune responses and immunotherapy effects in LGG patients.
To further analyze whether the genes in the ceRNA network are suitable for LGG and could become potential therapeutic targets for LGG, we analyzed LGG and adjacent tissues and defied DELGGmRNAs and DELGGlncRNAs. Next, we intersected these with the prognostic-related genes. A list of immune-related genes was then obtained from the ImmPort, and it was intersected with the list of identified mRNAs to identify potential immune-related genes. Finally, we identified the following 5 genes: HNF4G and ANGPTL2 (mRNAs), and CRNDE, HOTAIRM1, and GLYCTK-AS1 (lncRNAs). We then used these 5 genes to construct a new network (see Figure 5). Through verification with our LGG samples, we found that HNF4G, CRNDE, and HOTAIRM1 expression levels were downregulated in patients with IDH1 mutations (see Figure 6). However, their expression in glioma tissues was upregulated, which reflects the information found in TCGA and CGGA databases. Finally, we identified the CRNDE, HOTAIRM1/miRNA-206a/HNF4G axis, and found that the expression of HNF4G was positively correlated with CRNDE or HOTAIRM1 both in TCGA and CGGA databases.
The relationship between the expression of these genes with immune cell infiltration and PD-L1 expression was then analyzed. The expression levels of HNF4G and CRNDE were positively correlated to the infiltration of six types of immune cells (see Figure 10). The expression level of HOTAIRM1 was positively correlated to the infiltration of 5 types of immune cells except for CD4+ T cells. The expression levels of HNF4G, CRNDE, and HOTAIRM1 were also positively correlated to PD-L1 expression in TCGA database (see Figure 11). Consistent with TCGA, the expression levels of HNF4G were positively correlated to PD-L1 expression in the CGGA database; however, the expression levels of CRNDE and HOTAIRM1 were not statistically significantly correlated to PD-L1 expression in LGG (see Figure 11). Thus, HNF4G, CRNDE and HOTAIRMI may be closely related to immunity response.
HNF4G is a member of the orphan nuclear receptor superfamily (24). In the Chinese Han population, HNF4G polymorphisms are associated with ventilatory disease (25). Wang et al. (26) demonstrated that HNF4G acts as an oncogene and can promote the growth and metastasis of lung cancer cells. Further, HNF4G is also a prognostic factor for lung cancer. Sun et al. (27) found that miR-34 mediates the downregulation of HNF4G gene to inhibit bladder cancer cell growth and invasion. Tian et al. (28) demonstrated that HOTAIRM1/HOXA1 attenuates the immunosuppressive function of myeloid-derived suppressor cells in lung cancer, thereby promoting the immune response and delaying the progression of lung cancer. Lin et al. (29) found that HOTAIRM1 promotes cell growth and reduces apoptosis in glioma cell lines through the miR-873-5p/ZEB2 axis. Li et al. (30) suggested the formation of ceRNA network by CRNDE/MIR136-5P/Bcl-2 and Wnt2 that serves to regulate the biological characteristics of glioma. CRNDE can negatively regulate the miR-136-5P-mediated downregulation of Bcl-2 and Wnt2, thereby promoting the growth and metastasis of glioma cells (30). Previous studies have shown that the lncRNA CRNDE and HOTAIRM1 are closely associated with the prognosis of gliomas (31,32). There are many studies on CRNDE and HOTAIRM1 expression in gliomas, which indicates that these two lncRNAs are closely linked to the prognosis and biological behavior of gliomas, but there are no studies on HNF4G function in gliomas. The expression of HNF4G was positively correlated with CRNDE and HOTAIRM1 in TCGA and CGGA databases, and these genes may be related to each other. In addition, the three genes are closely related to tumor growth, prognosis, and the immune microenvironment and thus may serve as targets for treating LGG patients in the near future, especially those containing wild-type IDH1.
Our research had some limitations. First, while we identified the CRNDE, HOTAIRM1/miRNA-206a/HNF4G axis, these genes are positively correlated with each other, and thus their interrelationships need to be further investigated. Second, the sample size was small; thus, further research needs to be conducted with a larger sample size.
In summary, through our analysis, we constructed ceRNA networks with different IDH1 mutation statuses in LGG. By verification of the CGGA database and Quantitative Polymerase Chain Reaction (QT-PCR), we identified 1 mRNA and 2 lncRNAs that are related to immune cell infiltration and PD-L1 expression. These genes are related to LGG prognosis and IDH1 mutational status. We believe that HNF4G, CRNDE, and HOTAIRM1 play important roles in LGG, and these roles are related to IDH1 mutation status and the LGG immune microenvironment. Finally, by constructing a network of these genes, any competitive relationships affecting the development of LGG may be able to be identified.
Acknowledgments
We would like to thank Wei Shan for his assistance with the statistical analyses.
Funding: This study was supported by the Science and Education for Health Foundation of Suzhou for Youth (grant No. KJXW2019074); the Science and Technology Project Foundation of Suzhou (grant Nos. SS201651, SS201852, SS202093 and SYSD2020061); Project of science and technology development plan in Suzhou (grant Nos. SYSD2018138, SYSD202006) and the Jiangsu Province Medical key discipline (grant No. ZDXKC2016007).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6983/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6983/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6983/coif). All authors report funding from the Science and Education for Health Foundation of Suzhou for Youth (grant No. KJXW2019074); the Science and Technology Project Foundation of Suzhou (grant Nos. SS201651, SS201852 and SS202093); Project of science and technology development plan in Suzhou (grant Nos. SYSD2018138, SYSD202006) and the Jiangsu Province Medical key discipline (grant No. ZDXKC2016007). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by ethics board of the Affiliated Suzhou Hospital of Nanjing Medical University (No. KL901199) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet 2012;205:613-21. [Crossref] [PubMed]
- Buckner J, Giannini C, Eckel-Passow J, et al. Management of diffuse low-grade gliomas in adults - use of molecular diagnostics. Nat Rev Neurol 2017;13:340-51. [Crossref] [PubMed]
- Wesseling P, Capper D. WHO 2016 Classification of gliomas. Neuropathol Appl Neurobiol 2018;44:139-50. [Crossref] [PubMed]
- Kloosterhof NK, Bralten LB, Dubbink HJ, et al. Isocitrate dehydrogenase-1 mutations: a fundamentally new understanding of diffuse glioma? Lancet Oncol 2011;12:83-91. [Crossref] [PubMed]
- SongTao Q. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci 2012;103:269-73. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 2015;372:2481-98. [Crossref] [PubMed]
- Reuss DE, Kratz A, Sahm F, et al. Adult IDH wild type astrocytomas biologically and clinically resolve into other tumor entities. Acta Neuropathol 2015;130:407-17. [Crossref] [PubMed]
- Bao Z, Chen K, Krepel S, et al. High Glucose Promotes Human Glioblastoma Cell Growth by Increasing the Expression and Function of Chemoattractant and Growth Factor Receptors. Transl Oncol 2019;12:1155-63. [Crossref] [PubMed]
- Berghoff AS, Kiesel B, Widhalm G, et al. Correlation of immune phenotype with IDH mutation in diffuse glioma. Neuro Oncol 2017;19:1460-8. [Crossref] [PubMed]
- Kohanbash G, Carrera DA, Shrivastav S, et al. Isocitrate dehydrogenase mutations suppress STAT1 and CD8+ T cell accumulation in gliomas. J Clin Invest 2017;127:1425-37. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther 2016;1:15004. [Crossref] [PubMed]
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014;505:344-52. [Crossref] [PubMed]
- Wang JJ, Huang YQ, Song W, et al. Comprehensive analysis of the lncRNA-associated competing endogenous RNA network in breast cancer. Oncol Rep 2019;42:2572-82. [Crossref] [PubMed]
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015;43:D146-52. [Crossref] [PubMed]
- Chou CH, Chang NW, Shrestha S, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res 2016;44:D239-47. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Otasek D, Morris JH, Bouças J, et al. Cytoscape Automation: empowering workflow-based network analysis. Genome Biol 2019;20:185. [Crossref] [PubMed]
- Bhattacharya S, Andorf S, Gomes L, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res 2014;58:234-9. [Crossref] [PubMed]
- Cheng W, Ren X, Zhang C, et al. Gene Expression Profiling Stratifies IDH1-Mutant Glioma with Distinct Prognoses. Mol Neurobiol 2017;54:5996-6005. [Crossref] [PubMed]
- Garber ST, Hashimoto Y, Weathers SP, et al. Immune checkpoint blockade as a potential therapeutic target: surveying CNS malignancies. Neuro Oncol 2016;18:1357-66. [Crossref] [PubMed]
- Amankulor NM, Kim Y, Arora S, et al. Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes Dev 2017;31:774-86. [Crossref] [PubMed]
- Zhang X, Rao A, Sette P, et al. IDH mutant gliomas escape natural killer cell immune surveillance by downregulation of NKG2D ligand expression. Neuro Oncol 2016;18:1402-12. [Crossref] [PubMed]
- Bertrand S, Brunet FG, Escriva H, et al. Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Mol Biol Evol 2004;21:1923-37. [Crossref] [PubMed]
- Chen BD, Chen XC, Pan S, et al. TT genotype of rs2941484 in the human HNF4G gene is associated with hyperuricemia in Chinese Han men. Oncotarget 2017;8:26918-26. [Crossref] [PubMed]
- Wang J, Zhang J, Xu L, et al. Expression of HNF4G and its potential functions in lung cancer. Oncotarget 2018;9:18018-28. [Crossref] [PubMed]
- Sun H, Tian J, Xian W, et al. miR-34a inhibits proliferation and invasion of bladder cancer cells by targeting orphan nuclear receptor HNF4G. Dis Markers 2015;2015:879254. [Crossref] [PubMed]
- Tian X, Ma J, Wang T, et al. Long Non-Coding RNA HOXA Transcript Antisense RNA Myeloid-Specific 1-HOXA1 Axis Downregulates the Immunosuppressive Activity of Myeloid-Derived Suppressor Cells in Lung Cancer. Front Immunol 2018;9:473. [Crossref] [PubMed]
- Lin YH, Guo L, Yan F, et al. Long non-coding RNA HOTAIRM1 promotes proliferation and inhibits apoptosis of glioma cells by regulating the miR-873-5p/ZEB2 axis. Chin Med J (Engl) 2020;133:174-82. [Crossref] [PubMed]
- Li DX, Fei XR, Dong YF, et al. The long non-coding RNA CRNDE acts as a ceRNA and promotes glioma malignancy by preventing miR-136-5p-mediated downregulation of Bcl-2 and Wnt2. Oncotarget 2017;8:88163-78. [Crossref] [PubMed]
- Kiang KM, Zhang XQ, Zhang GP, et al. CRNDE Expression Positively Correlates with EGFR Activation and Modulates Glioma Cell Growth. Target Oncol 2017;12:353-63. [Crossref] [PubMed]
- Liang Q, Li X, Guan G, et al. Long non-coding RNA, HOTAIRM1, promotes glioma malignancy by forming a ceRNA network. Aging (Albany NY) 2019;11:6805-38. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)