
Overexpression of miR-328-5p influences cell growth and migration to promote NSCLC progression by targeting LOXL4
Introduction
With a high incidence and poor survival rates, lung cancer has been the leading cause of cancer-associated mortality worldwide. More than 80% of lung cancers are classified as non-small cell lung cancer (NSCLC). NSCLC can be divided into 2 major categories, namely squamous cell carcinoma and adenocarcinoma (1,2). Although chemotherapy, molecular targeted therapy, and immunotherapy have made promising advances in NSCLC treatment, the overall clinical outcomes are poor, with unfavorable prognosis and increased risk of treatment-related complications (3,4). In China, compared to most countries, the incidence and mortality of NSCLC are relatively high, causing enormous harm to human health (2,5), and poor areas still account for the dominant proportion of NSCLC cases in China (6). It is projected that the rate of NSCLC deaths in China may increase to approximately 40% in 2030 (7). Thus, there is an urgent need to find novel diagnostic and prognostic biomarkers for NSCLC.
MicroRNAs (miRNAs) are endogenous, highly conserved, single-stranded, non-coding small RNAs 18–23 nucleotides in length (8). They can lead to various diseases by binding specifically to the 3'-non-coding region of the antisense strand of the target gene (9). Increasing evidence has shown that miRNAs play an important role in regulating the progression of NSCLC (10-12). MiR-328 has been reported to function as a cancer-related miRNA. MiR-328 could increase reactive oxygen species (ROS) production and promote the adhesion and migration ability of monocytes during monocyte maturation (13). In lung cancer, miR-328 expression was significantly elevated and could promote cell migration (14-16).
In the present study, we found that the expression of miR-328-5p was upregulated in primary tumor tissues from NSCLC patients compared with adjacent tissues, and overexpression of miR-328-5p promoted NSCLC cell proliferation and migration. These results suggested that miR-328-5p may be a tumor promoter in NSCLC. We also demonstrated that miR-328-5p could directly bind to lysyl oxidase like 4 (LOXL4). This suggests that miR-328-5p may promote NSCLC cell proliferation and migration through targeted inhibition of LOXL4 expression. Taken together, these findings revealed that miR-328-5p/LOXL4 play a crucial role in NSCLC progression and highlight that miR-328-5p could be a potential biomarker for the diagnosis of NSCLC. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-345/rc).
Methods
Patients and tissue collection
Thirty paraffin-embedded lung cancer tissues and 30 pair-matched adjacent normal tissues were collected, along with the clinical data of the patients. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Nanfang Hospital and informed consent was taken from all the patients.
Total RNA isolation, miRNA isolation, and qRT-PCR
MiRNA was extracted from paraffin-embedded tissues using a Rapid microRNA Extraction Kit (centrifugal column type) (Beijing Baitaike Biotechnology Co., Ltd., China), and total RNA containing miRNA was extracted from cell lines using RNAiso Plus reagent (TaKaRa, Japan). The reverse transcription reaction was performed using the All-in-One™ First-Strand cDNA Synthesis Kit (GeneCopoeiaTM, Guangzhou, China). We performed real-time quantitative polymerase chain reaction (qRT-PCR) on a 7500 Real-time PCR System (Applied Biosystems, Foster City, CA, USA) using the All-in-One™ miRNA qRT-PCR Detection Kit (GeneCopoeiaTM) following the manufacturer’s protocol. The primers for miR-328-5p were designed by GeneCopoeiaTM. MiRNA expression was normalized to U6. Each sample was tested in triplicate.
Culture of lung cancer cells
Human A549 and SPC-A1 cells were maintained in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/mL penicillin, and 100 mg/mL streptomycin at 37 ℃ with 5% CO2.
MiR-328-5p modification in lung cancer cells
Lentiviral vectors were used to establish stable overexpression (LV-328-5p-m), inhibition (LV-328-5p-i), and negative control (LV-NC) NSCLC cells (obtained from GenePharma, Shanghai, China). These stable NSCLC cells expressed green fluorescent protein (GFP) and puromycin resistance gene to select stable cells for further analysis.
CCK8 and cell colony formation assays
Cells were seeded into 96-well plates at a density of 5×104/mL. Then, 10 µL of CCK8 solution (Donjindo, Japan) was added per well at the appointed time points. Absorbance at 450 nm was measured after a 2 h incubation. For cell colony formation assays, 500 cells were seeded in a 6-well plate and cultured for 2 weeks. The numbers of colonies per well were counted after staining with crystal violet. All studies were conducted with 3 replicates.
Cell cycle and apoptosis assays
For cell cycle analysis, NSCLC cells were plated into 6 well plates at 3×105 per well. After 24 h, cells were washed with phosphate-buffered saline (PBS) twice and trypsinized to collect cells, then cells were resuspended in cold 70% ethanol overnight. After treatment with RNase and staining with propidium iodide (PI), cell cycle position was analyzed by flow cytometry. For apoptosis assays, NSCLC cells were plated at a density of 3×105 per 6-well plate. After 24 h, cells were washed, collected, and stained with FITC-Annexin V, and cell apoptosis stages were analyzed by flow cytometry.
In vitro wound healing and migration assays
Cells (4×105) were seeded into 6-well plates. When the cells reached 95% confluence, wound healing assays were carried out with a sterile 10 µL pipette tip to straightly scratch 3 lines through the confluent monolayer. The cells were cultured with fresh medium, and wound closure was photographed at 24, 48, and 72 h. For the migration assay, cells (5×104) in serum-free medium were seeded into the upper chamber of a Transwell (Merck Millipore, USA), and after 48 h, the cells in the upper chamber were removed. The number of cells that adhered to the lower membrane was imaged after staining with 0.1% crystal violet. All studies were conducted with 3 replicates.
Establishment of tumor xenografts in mice
Male 3-week-old nude mice were purchased from Guangdong Medical Laboratory Animal Center (Guangzhou, China). After acclimation for a week in individually ventilated cages (IVC), 3×106 cells were subcutaneously injected into each flank (groups of LV-328-5p-m, LV-328-5p-i and LV-NC in 1 mouse). When the largest subcutaneous tumor of the mouse reached a volume of approximately 1,000 mm3, In-Vivo FX Pro (BRUKER, Germany) was applied to detect the fluorescence of subcutaneous tumors in mice. Subcutaneous tumors were resected following euthanasia, and size was calculated by tumor volume (mm3) = (L×W2)/2. Tumor weight was recorded. This experiment was conducted in the Guangzhou Institute of Sport Science and approved by the Standards for Animal Ethics in the Guangzhou Institute of Sport Science (GZTKSGNX-2015-2) and performed in accordance with the relevant experimental animal guidelines and regulations for the care and use of animals.
MiRNA profiling and RNA deep sequencing
Candidate target genes of miR-183-5p were detected by RNA deep sequencing. Total RNA was extracted from miR-183-5p stable NSCLC cells by Trizol reagent and the RNA-seq libraries were constructed by the TruSeq RNA Library Prep Kit (Illumina). RNA-seq experiments were performed by Novel Bioinformatics Co., Ltd. (Shanghai, China, http://www.novelbio.com/). Data analysis was performed using NovelBio Bio-Pharm Technology for differential mRNA expression in both LV-328-5p-m and LV-328-5p-i cell lines compared with LV-NC cells.
Analysis of pathways and Gene Ontology (GO) categories
To investigate the biological roles of differentially expressed miRNA target genes, biological process GO enrichment analysis based on the GO database was performed. Kyoto Encyclopedia of Genes and Genomes (KEGG) database analysis was performed for further assessment of the signaling pathways. The interactions between miRNAs and the candidate genes were visualized by the Cytoscape open source bioinformatics software platform. Functional interactions between genes of the carcinogenesis module were annotated by the STRING database. The significant GO categories were determined by Fisher’s exact test and P values were corrected by false discovery rate (FDR).
Luciferase reporter assay
For the reporter assays, miR-328-5p mimics along with pSI-check2-LOXL4-3'UTR were transfected into HEK293T cells. Lipofectamine 2000 transfection reagent (Invitrogen) was used in this process. Luciferase activity was evaluated using a dual-luciferase reporter assay system (Promega), according to the manufacturer’s instructions. Firefly luciferase activity was used for normalization.
Western blot analysis
Stably expressed NSCLC cells with or without miR-328-5p expression were collected, washed with PBS, and lysed with RIPA buffer. Equal amounts of cell lysates (20 µg) were separated by SDS-PAGE and transferred onto PVDF (Millipore, Billerica, MA, USA) membranes. Rabbit polyclonal anti-LOXL4 (1:1,000, bs-5790R, Bioss) and mouse polyclonal anti-GAPDH (1:1,000, #5174, Cell Signaling Technology) antibodies were added to the membranes and incubated overnight at 4 ℃. The anti-GAPDH antibody was used as the internal reference. The bands were exposed by ECL reagent (Thermo Fisher Scientific Inc., MA, USA) and analyzed by ImageJ software (NIH, Bethesda, MD, USA).
Statistical analysis
Statistical analysis was performed using SPSS (v. 22.0, Chicago, IL, USA). All data were presented as the mean ± SEM. The two-tailed Student’s t-test or analysis of variance (ANOVA) were used to compare unpaired samples. The paired Student’s t-test was used to compare paired groups of samples. The statistical significance of all tests was defined as P<0.05.
Results
Expression of miR-328-5p in lung cancer tissue and its correlation with the clinicopathological features of patients
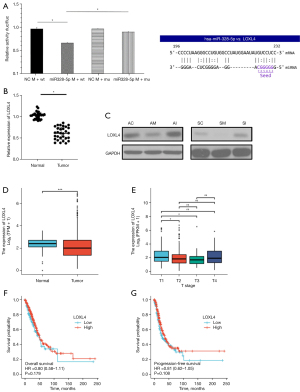
The miR-328-5p expression level was measured by qRT-PCR in 30 NSCLC and pair-matched adjacent normal lung tissue specimens to determine whether miR-328-5p was upregulated in NSCLC. The results showed that miR-328-5p expression was significantly increased (P<0.05) in NSCLC tissues relative to their matched controls (Figure 1A). Then, we used The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov/) to profile the expression of miR-328-5p in NSCLC tumor and adjacent normal tissues, which confirmed that miR-328-5p was significantly overexpressed in NSCLC samples (Figure 1B). Next, the correlation between miR-328-5p expression and clinicopathological parameters were assessed. Results showed that miR-328-5p expression levels in NSCLC were significantly associated with histological tumor type (P<0.05) and differentiation (P<0.05), but not with age, gender, tumor size, and TNM stage (Table 1). These results demonstrated that the over-expression of miR-328-5p might play an important role in lung cancer progression and development.
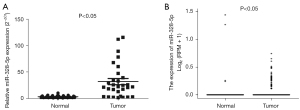
Table 1
| Variable | n | MiR-328-5p expression | P value | |
|---|---|---|---|---|
| Low | High | |||
| Gender | ||||
| Male | 21 | 6 | 15 | >0.05 |
| Female | 9 | 3 | 6 | |
| Age, years | ||||
| ≥50 | 17 | 4 | 13 | >0.05 |
| <50 | 13 | 3 | 10 | |
| Tumor volume, mm3 | ||||
| ≥5 | 9 | 2 | 7 | >0.05 |
| <5 | 21 | 5 | 16 | |
| Pathological type | ||||
| Adenocarcinoma | 18 | 3 | 15 | *<0.05 |
| Squamous cell carcinoma | 12 | 4 | 8 | |
| Differentiation degree | ||||
| High | 3 | 2 | 1 | *<0.05 |
| Medium + low | 27 | 7 | 20 | |
| TNM stage | ||||
| I–II | 20 | 5 | 15 | >0.05 |
| III–IV | 10 | 3 | 7 | |
Low/high by the mean expression of miR-328-5p. Pearson χ2 test. *, P<0.05 was considered statistically significant. NSCLC, non-small cell lung cancer.
Transfection of miR-328-5p in NSCLC cell lines
In terms of the correlation between miR-328-5p expression and NSCLC progression, we further investigated the effect of miR-328-5p on the proliferation and migration abilities of NSCLC cells. NSCLC cell lines were transfected with LV-328-5p-m, LV-328-5p-I, or LV-NC to establish the stable miR-328-5p overexpression or knockdown NSCLC cell lines. Then, the expression levels of miR-328-5p in cells were quantified via qRT-PCR. Compared with the expression of LV-NC cells, miR-328-5p was significantly upregulated in LV-328-5p-m transfected NSCLC cells, while LV-328-5p-i downregulated the expression of miR-328-5p (Figure 2A).
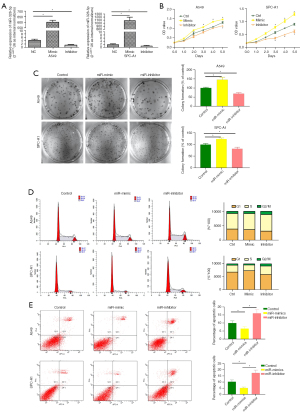
Overexpression of miR-328-5p promotes the proliferation of NSCLC cells
The role of miR-328-5p in NSCLC cell (A549 and SPC-A1) proliferation was examined by the CCK8 and colony formation assays. Results of the CCK8 assay demonstrated that overexpressed miR-328-5p significantly promoted A549 (P<0.05) and SPC-A-1 (P<0.05) cell proliferation, while downregulation of miR-328-5p reversed the inhibitory role of miR-328-5p on A549 and SPC-A-1 cell proliferation (Figure 2B). These results were confirmed by the colony formation assay, which was used to investigate the role of miR-328-5p on NSCLC cell clonogenic survival (Figure 2C). Moreover, flow cytometry analysis revealed that overexpressing miR-328-5p could significantly promote cell cycle arrest at the G2/M phase and reduce apoptosis in NSCLC cells (Figure 2D,2E).
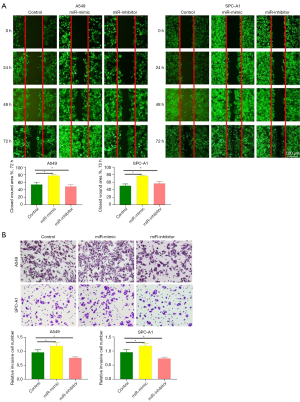
Overexpression of miR-328-5p promotes the migration of NSCLC cells
Considering the correlation between the expression of miR-328-5p and the progression of NSCLC, we used wound healing assays and Transwell assays in A549 and SPC-A1 cell lines transfected with either LV-328-5p-m, LV-328-5p-i, or LV-NC. As shown in Figure 3A,3B, compared with LV-NC cells, the wound healing ability and migration ability of stable miR-328-5p overexpressed cells were significantly increased. However, wound healing and migration were inhibited in NSCLC cells transfected with LV-328-5p-i compared with LV-NC cells. These results suggest that miR-183-5p can promote the migratory capacity of NSCLC cells and accelerate NSCLC progression.
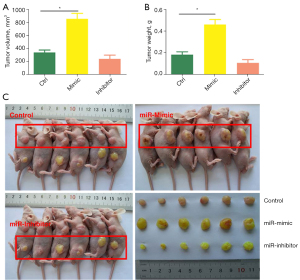
MiR-328-5p promotes the tumor growth of NSCLC cells in vivo
To verify whether miR-328-5p could promote NSCLC tumor growth in vivo, nude mice received subcutaneous injections of SPC-A1 cells transfected with either LV-328-5p-m, LV-328-5p-i, or LV-NC. After 28 days, the tumor volume and weight of mice injected with LV-328-5p-m SPC-A1 cells evidently increased compared with the LV-NC group (Figure 4A,4B). Additionally, the mice injected with LV-328-5p-i SPC-A1 cells displayed significantly decreased tumor volume and weight compared with the mice injected with LV-328-5p-m SPC-A1 cells (Figure 4C). Consistent with the in vitro data of the present study, these data indicated that miR-328-5p promotes the tumor growth of NSCLC cells in vivo.
LOXL4 is directly negatively regulated by miR-328-5p in NSCLC cells
To further clarify the molecular mechanisms of miR-328-5p in tumorigenesis, we overexpressed miR-328-5p and screened the downregulated differentially expressed target genes by RNA deep sequencing in NSCLC cells. A total of 14 downregulated genes were found in miR-328-5p overexpressed cells (Figure 5A). Notably, the KEGG pathways included channel activity, passive transmembrane transporter activity, ion channel complex, and copper ion binding, among others (Figure 5B-5E) (Table 2). Among the genes with several functions, we were interested in whether LOXL4, a member of the copper-dependent enzyme family, could be downregulated by miR-328-5p in NSCLC. To determine the correlation between miR-328-5p and LOXL4, luciferase reporter assays were used to determine whether miR-328-5p could recognize the 3'UTR of LOXL4 and regulate the expression of LOXL4 by their combined putative sites. Results showed that LOXL4 was able to match the intracellular miR-328-5p seed sequence at the 3'UTR region and the luciferase activity was significantly decreased with miR-328-5p overexpression, further indicating that LOXL4 was the direct target gene of miR-328-5p (Figure 6A). A qRT-PCR assay was used to measure the expression of LOXL4 in NSCLC tissues, and the results showed that LOXL4 was obviously downregulated in both NSCLC and normal tissues (Figure 6B). Then, a western blot assay was used to evaluate the regulatory effect of miR-328-5p on LOXL4 expression in NSCLC cells. Results revealed that overexpression of miR-328-5p decreased the protein levels of LOXL4 (Figure 6C). Using UCSC XENA (https://xenabrowser.net/datapages/) profiling of LOXL4 expression in NSCLC tumor tissues and normal tissues, we confirmed that LOXL4 was significantly decreased in NSCLC samples. Moreover, statistical correlation analysis showed significant negative regulation between LOXL4 expression and TNM stage (TCGA, https://portal.gdc.cancer.gov/). Overall survival and progression-free survival were lower in NSCLC with downregulation of LOXL4, but showed no significant difference (Figure 6D-6G). These data indicate that overexpressed miR-328-5p in NSCLC interacted with LOXL4.
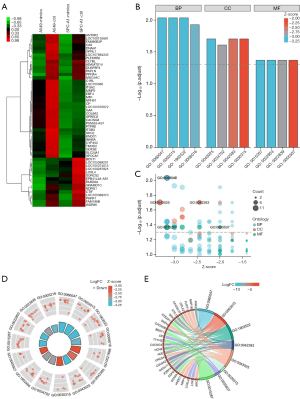
Table 2
| Ontology | ID | Description | Padjust | Gene ID |
|---|---|---|---|---|
| BP | GO:0060047 | Heart contraction | 0.0092 | ANK2/CACNA1F/STC1/SPX/CACNG6/CACNG4/HCN4/ACE/KCNJ11/P2RX4 |
| BP | GO:0003015 | Heart process | 0.0092 | ANK2/CACNA1F/STC1/SPX/CACNG6/CACNG4/HCN4/ACE/KCNJ11/P2RX4 |
| BP | GO:1903522 | Regulation of blood circulation | 0.0092 | ANK2/CACNA1F/STC1/SPX/CACNG6/CACNG4/HCN4/ACE/KCNJ11/P2RX4 |
| BP | GO:0008016 | Regulation of heart contraction | 0.0118 | ANK2/CACNA1F/STC1/SPX/CACNG6/CACNG4/HCN4/KCNJ11/P2RX4 |
| BP | GO:0021696 | Cerebellar cortex morphogenesis | 0.0124 | GLI1/SERPINE2/WNT7A/KNDC1 |
| BP | GO:0035637 | Multicellular organismal signaling | 0.0124 | ANK2/CACNA1F/CACNG6/CACNG4/HCN4/KCNJ11/P2RX4/SCN4A |
| CC | GO:0042383 | Sarcolemma | 0.0198 | ANK2/CACNG6/CACNG4/COL6A2/KCNJ11/CDH2 |
| CC | GO:0043025 | Neuronal cell body | 0.0198 | MAP1B/FLRT1/CPLX2/CACNA1F/SNPH/UNC5A/KCNJ11/P2RX4/KNDC1/NRP1/ARC |
| CC | GO:0030315 | T-tubule | 0.0198 | ANK2/CACNG6/CACNG4/KCNJ11 |
| BP | GO:0021681 | Cerebellar granular layer development | 0.0199 | SERPINE2/WNT7A/KNDC1 |
| BP | GO:0021587 | Cerebellum morphogenesis | 0.0209 | GLI1/SERPINE2/WNT7A/KNDC1 |
| CC | GO:0034702 | Ion channel complex | 0.0246 | CACNA1F/CACNG6/BEST1/TTYH2/CACNG4/HCN4/KCNJ11/SCN4A |
| BP | GO:0021575 | Hindbrain morphogenesis | 0.0269 | GLI1/SERPINE2/WNT7A/KNDC1 |
| BP | GO:1900451 | Positive regulation of glutamate receptor signaling pathway | 0.0269 | NECAB2/CACNG4/ARC |
| CC | GO:1902495 | Transmembrane transporter complex | 0.0312 | CACNA1F/CACNG6/BEST1/TTYH2/CACNG4/HCN4/KCNJ11/SCN4A |
| CC | GO:1990351 | Transporter complex | 0.0312 | CACNA1F/CACNG6/BEST1/TTYH2/CACNG4/HCN4/KCNJ11/SCN4A |
| CC | GO:0097060 | Synaptic membrane | 0.0354 | ANK2/SNPH/CACNG4/DLG5/CDH2/DNAJA3/GRM4/NRP1/ARC |
| BP | GO:0021695 | Cerebellar cortex development | 0.0361 | GLI1/SERPINE2/WNT7A/KNDC1 |
| BP | GO:0051899 | Membrane depolarization | 0.0361 | ANK2/CACNG4/HCN4/P2RX4/SCN4A |
| BP | GO:0043062 | Extracellular structure organization | 0.0361 | RAMP2/ITGAM/LPL/ADAMTS14/NPHS1/ECM2/COL6A2/THSD4/LOXL4/LOX |
| BP | GO:0051047 | Positive regulation of secretion | 0.0361 | CD33/ITGAM/LPL/TNFRSF14/OPRL1/INHBA/CD177/EXPH5/TWIST1/P2RX4 |
| BP | GO:0061337 | Cardiac conduction | 0.0361 | ANK2/CACNA1F/CACNG6/CACNG4/HCN4/KCNJ11 |
| BP | GO:0006816 | Calcium ion transport | 0.0361 | CD33/ANK2/RAMP2/CACNA1F/STC1/OPRL1/CACNG6/BEST1/CACNG4/P2RX4 |
| BP | GO:0050804 | Modulation of chemical synaptic transmission | 0.0361 | MAP1B/CPLX2/CACNG4/SERPINE2/CDH2/WNT7A/GRM4/EGR2/MCTP1/ARC |
| BP | GO:0099177 | Regulation of trans-synaptic signaling | 0.0361 | MAP1B/CPLX2/CACNG4/SERPINE2/CDH2/WNT7A/GRM4/EGR2/MCTP1/ARC |
| BP | GO:0006942 | Regulation of striated muscle contraction | 0.0361 | ANK2/STC1/HCN4/P2RX4/SCN4A |
| BP | GO:0044342 | Type B pancreatic cell proliferation | 0.0361 | NUPR1/IGFBP3/SGPP2 |
| BP | GO:0045216 | Cell-cell junction organization | 0.0397 | ANK2/RAMP2/CD177/ACE/DLG5/CDH2 |
| BP | GO:0030198 | Extracellular matrix organization | 0.0397 | RAMP2/ITGAM/ADAMTS14/NPHS1/ECM2/COL6A2/THSD4/LOXL4/LOX |
| BP | GO:0044062 | Regulation of excretion | 0.0409 | STC1/SPX/NPHS1 |
| MF | GO:0005507 | Copper ion binding | 0.0428 | CP/P2RX4/LOXL4/LOX |
| MF | GO:0015267 | Channel activity | 0.0428 | CACNA1F/STEAP1/CACNG6/BEST1/TTYH2/CACNG4/HCN4/KCNJ11/P2RX4/SCN4A |
| MF | GO:0022803 | Passive transmembrane transporter activity | 0.0428 | CACNA1F/STEAP1/CACNG6/BEST1/TTYH2/CACNG4/HCN4/KCNJ11/P2RX4/SCN4A |
| MF | GO:0022839 | Ion gated channel activity | 0.0428 | CACNA1F/CACNG6/TTYH2/CACNG4/HCN4/KCNJ11/P2RX4/SCN4A |
| MF | GO:0005216 | Ion channel activity | 0.0428 | CACNA1F/CACNG6/BEST1/TTYH2/CACNG4/HCN4/KCNJ11/P2RX4/SCN4A |
| MF | GO:0022836 | Gated channel activity | 0.0428 | CACNA1F/CACNG6/TTYH2/CACNG4/HCN4/KCNJ11/P2RX4/SCN4A |
| MF | GO:0022838 | Substrate-specific channel activity | 0.0428 | CACNA1F/CACNG6/BEST1/TTYH2/CACNG4/HCN4/KCNJ11/P2RX4/SCN4A |
| MF | GO:0005244 | Voltage-gated ion channel activity | 0.0428 | CACNA1F/CACNG6/CACNG4/HCN4/KCNJ11/SCN4A |
| MF | GO:0022832 | Voltage-gated channel activity | 0.0428 | CACNA1F/CACNG6/CACNG4/HCN4/KCNJ11/SCN4A |
| BP | GO:0016358 | Dendrite development | 0.0505 | MAP1B/FLRT1/DLG5/WNT7A/KNDC1/NRP1/ARC |
| MF | GO:0022843 | Voltage-gated cation channel activity | 0.0518 | CACNA1F/CACNG6/CACNG4/HCN4/KCNJ11 |
| CC | GO:0034703 | Cation channel complex | 0.0523 | CACNA1F/CACNG6/CACNG4/HCN4/KCNJ11/SCN4A |
| CC | GO:0005891 | Voltage-gated calcium channel complex | 0.0523 | CACNA1F/CACNG6/CACNG4 |
| CC | GO:1990454 | L-type voltage-gated calcium channel complex | 0.0523 | CACNG6/CACNG4 |
| BP | GO:0006937 | Regulation of muscle contraction | 0.0551 | ANK2/STC1/SPX/HCN4/P2RX4/SCN4A |
| BP | GO:0034765 | Regulation of ion transmembrane transport | 0.0564 | ANK2/CACNA1F/OPRL1/CACNG6/CACNG4/HCN4/KCNJ11/TWIST1/SCN4A/ARC |
| BP | GO:0070838 | Divalent metal ion transport | 0.0564 | CD33/ANK2/RAMP2/CACNA1F/STC1/OPRL1/CACNG6/BEST1/CACNG4/P2RX4 |
| BP | GO:0021602 | Cranial nerve morphogenesis | 0.0577 | GLI3/NRP1/EGR2 |
| BP | GO:0072511 | Divalent inorganic cation transport | 0.0577 | CD33/ANK2/RAMP2/CACNA1F/STC1/OPRL1/CACNG6/BEST1/CACNG4/P2RX4 |
| BP | GO:0050808 | Synapse organization | 0.0587 | ITGAM/MAP1B/FLRT1/DLG5/CDH2/DNAJA3/WNT7A/NRP1/ARC |
| BP | GO:0008217 | Regulation of blood pressure | 0.0587 | RAMP2/SPX/OPRL1/ACE/P2RX4/NPR3 |
| BP | GO:0021953 | Central nervous system neuron differentiation | 0.0587 | INHBA/GLI3/CHD5/WNT7A/KNDC1/NRP1 |
| BP | GO:0051966 | Regulation of synaptic transmission, glutamatergic | 0.0587 | CACNG4/SERPINE2/CDH2/GRM4 |
| BP | GO:2001259 | Positive regulation of cation channel activity | 0.0587 | ANK2/CACNG4/KCNJ11/ARC |
| BP | GO:0007043 | Cell-cell junction assembly | 0.0610 | ANK2/RAMP2/ACE/DLG5/CDH2 |
| BP | GO:0048167 | Regulation of synaptic plasticity | 0.0610 | MAP1B/CPLX2/SERPINE2/EGR2/MCTP1/ARC |
| CC | GO:0045211 | Postsynaptic membrane | 0.0636 | ANK2/CACNG4/DLG5/CDH2/DNAJA3/NRP1/ARC |
| CC | GO:0014704 | Intercalated disc | 0.0636 | ANK2/KCNJ11/CDH2 |
| MF | GO:0031404 | Chloride ion binding | 0.0646 | ACE/NPR3 |
| MF | GO:0005245 | Voltage-gated calcium channel activity | 0.0646 | CACNA1F/CACNG6/CACNG4 |
| MF | GO:0017046 | Peptide hormone binding | 0.0646 | RAMP2/INHBA/NPR3 |
| MF | GO:0005261 | Cation channel activity | 0.0646 | CACNA1F/CACNG6/CACNG4/HCN4/KCNJ11/P2RX4/SCN4A |
| MF | GO:0042562 | Hormone binding | 0.0646 | RAMP2/INHBA/ALDH1A1/NPR3 |
| MF | GO:0004029 | Aldehyde dehydrogenase (NAD) activity | 0.0646 | ALDH1L2/ALDH1A1 |
| MF | GO:0008201 | Heparin binding | 0.0646 | LPL/LTBP2/ECM2/SERPINE2/NRP1 |
| MF | GO:0016641 | Oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor | 0.0661 | LOXL4/LOX |
| BP | GO:0003002 | Regionalization | 0.0765 | ITGAM/NBL1/GLI1/GLI3/WNT7A/NRP1/EGR2/ARC |
| BP | GO:0008589 | Regulation of smoothened signaling pathway | 0.0765 | GLI1/SERPINE2/GLI3/DLG5 |
| BP | GO:0050866 | Negative regulation of cell activation | 0.0765 | IL13RA2/TNFRSF14/INHBA/SERPINE2/GLI3/DLG5 |
| BP | GO:0032355 | Response to estradiol | 0.0765 | RAMP2/MAP1B/OPRL1/KCNJ11/WNT7A |
| BP | GO:0021675 | Nerve development | 0.0765 | SERPINE2/GLI3/NRP1/EGR2 |
| BP | GO:0086010 | Membrane depolarization during action potential | 0.0796 | ANK2/HCN4/SCN4A |
| BP | GO:0007389 | Pattern specification process | 0.0796 | ITGAM/NBL1/STC1/GLI1/GLI3/WNT7A/NRP1/EGR2/ARC |
| BP | GO:0055117 | Regulation of cardiac muscle contraction | 0.0819 | ANK2/STC1/HCN4/P2RX4 |
| BP | GO:0086070 | SA node cell to atrial cardiac muscle cell communication | 0.0835 | ANK2/HCN4 |
| BP | GO:0150063 | Visual system development | 0.0835 | ITGAM/INHBA/GLI3/TWIST1/CRYGN/WNT7A/CRYBA2/NRP1 |
| BP | GO:0048880 | Sensory system development | 0.0852 | ITGAM/INHBA/GLI3/TWIST1/CRYGN/WNT7A/CRYBA2/NRP1 |
| BP | GO:0034330 | Cell junction organization | 0.0852 | ANK2/RAMP2/CD177/ACE/DLG5/CDH2/NRP1 |
| BP | GO:0021561 | Facial nerve development | 0.0852 | NRP1/EGR2 |
| BP | GO:0021604 | Cranial nerve structural organization | 0.0852 | NRP1/EGR2 |
| BP | GO:0021610 | Facial nerve morphogenesis | 0.0852 | NRP1/EGR2 |
| BP | GO:0031915 | Positive regulation of synaptic plasticity | 0.0852 | MAP1B/CPLX2 |
| BP | GO:1902563 | Regulation of neutrophil activation | 0.0852 | ITGAM/CD177 |
| BP | GO:0017157 | Regulation of exocytosis | 0.0852 | IL13RA2/ITGAM/CPLX2/CD177/EXPH5/WNT7A |
| BP | GO:0007409 | Axonogenesis | 0.0852 | MAP1B/GLI3/CRMP1/UNC5A/EPHA10/CDH2/WNT7A/NRP1/EGR2 |
| BP | GO:0060021 | Roof of mouth development | 0.0852 | INHBA/GLI3/TWIST1/WNT7A |
| BP | GO:0007584 | Response to nutrient | 0.0852 | MAP1B/LPL/STC1/CYP24A1/KCNJ11/P2RX4 |
| BP | GO:0030900 | Forebrain development | 0.0852 | ITGAM/GLI1/INHBA/GLI3/CHD5/CDH2/WNT7A/NRP1 |
| BP | GO:0009953 | Dorsal/ventral pattern formation | 0.0860 | NBL1/GLI1/GLI3/WNT7A |
| BP | GO:0086004 | Regulation of cardiac muscle cell contraction | 0.0875 | ANK2/STC1/HCN4 |
| BP | GO:0030902 | Hindbrain development | 0.0875 | GLI1/SERPINE2/WNT7A/KNDC1/EGR2 |
| BP | GO:0060019 | Radial glial cell differentiation | 0.0892 | GLI3/CDH2 |
| BP | GO:0001649 | Osteoblast differentiation | 0.0894 | GLI1/GLI3/CYP24A1/TWIST1/IGFBP3/LOX |
| BP | GO:0035249 | Synaptic transmission, glutamatergic | 0.0894 | CACNG4/SERPINE2/CDH2/GRM4 |
| MF | GO:0005248 | Voltage-gated sodium channel activity | 0.0895 | HCN4/SCN4A |
| MF | GO:0016638 | Oxidoreductase activity, acting on the CH-NH2 group of donors | 0.0895 | LOXL4/LOX |
| MF | GO:0005179 | Hormone activity | 0.0895 | STC1/SPX/INHBA/INHBE |
| MF | GO:0005262 | Calcium channel activity | 0.0895 | CACNA1F/CACNG6/CACNG4/P2RX4 |
| BP | GO:0043271 | Negative regulation of ion transport | 0.0909 | CD33/STC1/OPRL1/SERPINE2/TWIST1 |
| BP | GO:0043300 | Regulation of leukocyte degranulation | 0.0909 | IL13RA2/ITGAM/CD177 |
| BP | GO:0001503 | Ossification | 0.0909 | STC1/GLI1/GLI3/CYP24A1/TWIST1/IGFBP3/EGR2/LOX |
| BP | GO:0003084 | Positive regulation of systemic arterial blood pressure | 0.0909 | SPX/ACE |
| BP | GO:0043116 | Negative regulation of vascular permeability | 0.0909 | RAMP2/MIR23A |
| BP | GO:0061029 | Eyelid development in camera-type eye | 0.0909 | INHBA/TWIST1 |
| BP | GO:0061469 | Regulation of type B pancreatic cell proliferation | 0.0909 | NUPR1/SGPP2 |
| BP | GO:1903532 | Positive regulation of secretion by cell | 0.0909 | CD33/ITGAM/LPL/TNFRSF14/INHBA/CD177/EXPH5/TWIST1 |
| BP | GO:1903305 | Regulation of regulated secretory pathway | 0.0909 | IL13RA2/ITGAM/CPLX2/CD177/WNT7A |
| BP | GO:0043010 | Camera-type eye development | 0.0909 | INHBA/GLI3/TWIST1/CRYGN/WNT7A/CRYBA2/NRP1 |
| BP | GO:1903115 | Regulation of actin filament-based movement | 0.0909 | ANK2/STC1/HCN4 |
| BP | GO:0099173 | Postsynapse organization | 0.0951 | CDH2/DNAJA3/WNT7A/NRP1/ARC |
| BP | GO:0007215 | Glutamate receptor signaling pathway | 0.0967 | NECAB2/CACNG4/GRM4/ARC |
| BP | GO:0035810 | Positive regulation of urine volume | 0.0970 | OPRL1/NPR3 |
| MF | GO:0019955 | Cytokine binding | 0.0982 | IL13RA2/NBL1/TNFRSF14/NRP1 |
| BP | GO:0034329 | Cell junction assembly | 0.0999 | ANK2/RAMP2/ACE/DLG5/CDH2/NRP1 |
| BP | GO:0021549 | Cerebellum development | 0.0999 | GLI1/SERPINE2/WNT7A/KNDC1 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; NSCLC, non-small cell lung cancer; BP, biological process; MF, molecular function; CC, cellular component.
Discussion
In this experimental and clinical study, we found that the progression of NSCLC is closely related to the expression of miR-328-5p. Overexpressed miR-328-5p could promote the proliferation and migration of NSCLC cells and promote tumor growth in nude mice. This indicated that high expression of miR-328-5p is a high-risk factor in NSCLC. Furthermore, according to miRNA deep sequencing and KEGG pathway/GO analysis categories, we found that overexpression of miR-328-5p inhibited LOXL4 expression and was significantly correlated with the proliferation and migration of NSCLC cells. These results showed that, in NSCLC, miR-328-5p is a novel tumor suppressor by targeting LOXL4.
With a high incidence and poor survival rates, NSCLC has been the leading cause of cancer-associated mortality worldwide (1,2,17). NSCLC can be divided into 2 major categories, namely squamous cell carcinoma and adenocarcinoma. Although chemotherapy, molecular targeted therapy, and immunotherapy have made promising advances in NSCLC treatment, the overall clinical outcomes are poor, with poor prognosis and increased risk of treatment-related complications (4,18,19). Thus, there is an urgent need to find novel diagnostic and prognostic biomarkers for NSCLC.
Serving as essential regulators in tumorigenesis and tumor development, miRNAs can regulate different tumor processes, including initiation, promotion, malignant conversion, progression, and metastasis (20,21). Aberrant miRNA expression levels are promising and novel makers for tumor diagnosis (22), prognosis (23), and treatment (24,25). Emerging evidence demonstrates that miRNAs play important roles in the pathogenesis of lung cancer (26-28). MiR-151a could promote cell proliferation and migration by regulating E-cadherin in lung cancer (29). Overexpression of miR-1275 could target HIF-1α and resulted in shorter overall survival and recurrence-free survival in NSCLC (27). In this study, we found that miR-328-5p was overexpressed in NSCLC, and was significantly associated with histological tumor type and differentiation. Additionally, overexpression of miR-328-5p significantly promoted cell proliferation and migration. These results suggest that miR-328-5p acts as a crucial tumor promoter in NSCLC carcinogenesis.
MiRNAs exert their numerous functional roles by their specific interactions with upregulated or downregulated mRNAs of target genes (30). In order to investigate the target gene regulated by miR-328-5p, we used high throughput sequencing to screen out the differentially expressed genes. Based on sequencing data for target genes in miR-328-5p overexpressed cells, we found that 11 genes were significantly downregulated. These potential target genes and the analysis of several key pathways showed that miR-328-5p may be involved in pathways such as channel activity, passive transmembrane transporter activity, ion channel complex, and copper ion binding, among others. Lysyl oxidases (LOX) are a family of copper-dependent enzymes comprised of 5 paralogues: the prototypical LOX and 4 LOX-like (LOXL) enzymes labeled 1 through 4 (31). The main function of the LOX family (LOX, LOXL1, LOXL2, LOXL3, and LOXL4) is to cross-link collagen and elastin, shaping the structure and strength of the extracellular matrix (ECM) (32). LOXL4 is widely expressed in vivo. So far, the role of LOXL4 in human malignancies is scarcely understood. Recent studies showed that LOXL4 has both tumor oncogenic and tumor suppressor roles (33,34). Researchers found that LOXL4 was significantly correlated with breast cancer (24), gastric cancer (35), and hepatocellular carcinoma (33), whereas it acts as a tumor suppressor in lung cancer (34) and bladder cancer (36). However, the roles and mechanisms of LOXL4 in NSCLC remain unclear. In the present study, we assessed the correlation between LOXL4 expression and the clinicopathological characteristics of NSCLC patients. We found significantly lower expression of LOXL4 in NSCLC tumor tissues than in normal tissues. Moreover, we found that LOXL4 expression was correlated with the TNM stage, but had a low correlation with overall survival and progression-free survival in NSCLC patients.
In summary, we found that miR-328-5p overexpression promoted cell proliferation and migration, and can be defined as a crucial tumor promoter by downregulating LOXL4 expression in NSCLC. These results suggest that miR-328-5p may serve as a predictive marker for prognosis, as well as a therapeutic target for NSCLC patients.
Acknowledgments
Funding: This work was supported by the National Science Foundation of China (Nos. 81673840, 81803877, 81873205); the Natural Science Foundation of Guangdong Province, China (No. 2020B1515120063); the Guangdong Basic and Applied Basic Research Foundation, China (No. 2020A1515110651); Social science and Technology Development Foundation of Dongguan (No. 202050715001207); Medical Scientific Research Foundation of Guangdong Province, China (No. 20201151015201); and the Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine (No. ZYYCXTD-C-202001).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-345/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-345/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-345/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Nanfang Hospital and informed consent was taken from all the patients. Animal experiment was approved by the Standards for Animal Ethics in the Guangzhou Institute of Sport Science (No. GZTKSGNX-2015-2) and performed in accordance with the relevant experimental animal guidelines and regulations for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Liu Y, Fan J, Xu T, et al. Extracellular vesicle tetraspanin-8 level predicts distant metastasis in non-small cell lung cancer after concurrent chemoradiation. Sci Adv 2020;6:eaaz6162. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Okuyama A. Lung cancer incidence rates in the world from the Cancer Incidence in Five Continents XI. Jpn J Clin Oncol 2018;48:300-1. [Crossref] [PubMed]
- Sullivan DR, Forsberg CW, Ganzini L, et al. Longitudinal Changes in Depression Symptoms and Survival Among Patients With Lung Cancer: A National Cohort Assessment. J Clin Oncol 2016;34:3984-91. [Crossref] [PubMed]
- Cao M, Chen W. Epidemiology of lung cancer in China. Thorac Cancer 2019;10:3-7. [Crossref] [PubMed]
- Feng RM, Zong YN, Cao SM, et al. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun (Lond) 2019;39:22. [Crossref] [PubMed]
- Martín-Sánchez JC, Lunet N, González-Marrón A, et al. Projections in Breast and Lung Cancer Mortality among Women: A Bayesian Analysis of 52 Countries Worldwide. Cancer Res 2018;78:4436-42. [Crossref] [PubMed]
- Zeng F, Wang Q, Wang S, et al. Linc00173 promotes chemoresistance and progression of small cell lung cancer by sponging miR-218 to regulate Etk expression. Oncogene 2020;39:293-307. [Crossref] [PubMed]
- Pan J, Fang S, Tian H, et al. lncRNA JPX/miR-33a-5p/Twist1 axis regulates tumorigenesis and metastasis of lung cancer by activating Wnt/β-catenin signaling. Mol Cancer 2020;19:9. [Crossref] [PubMed]
- Xu Y, Lin J, Jin Y, et al. The miRNA hsa-miR-6515-3p potentially contributes to lncRNA H19-mediated-lung cancer metastasis. J Cell Biochem 2019;120:17413-21. [Crossref] [PubMed]
- Zhang L, Lin J, Ye Y, et al. Serum MicroRNA-150 Predicts Prognosis for Early-Stage Non-Small Cell Lung Cancer and Promotes Tumor Cell Proliferation by Targeting Tumor Suppressor Gene SRCIN1. Clin Pharmacol Ther 2018;103:1061-73. [Crossref] [PubMed]
- Gallach S, Jantus-Lewintre E, Calabuig-Fariñas S, et al. MicroRNA profiling associated with non-small cell lung cancer: next generation sequencing detection, experimental validation, and prognostic value. Oncotarget 2017;8:56143-57. [Crossref] [PubMed]
- Saul MJ, Hegewald AB, Emmerich AC, et al. Mass Spectrometry-Based Proteomics Approach Characterizes the Dual Functionality of miR-328 in Monocytes. Front Pharmacol 2019;10:640. [Crossref] [PubMed]
- Delic S, Lottmann N, Stelzl A, et al. MiR-328 promotes glioma cell invasion via SFRP1-dependent Wnt-signaling activation. Neuro Oncol 2014;16:179-90. [Crossref] [PubMed]
- Yin Y, Castro AM, Hoekstra M, et al. Fibroblast Growth Factor 9 Regulation by MicroRNAs Controls Lung Development and Links DICER1 Loss to the Pathogenesis of Pleuropulmonary Blastoma. PLoS Genet 2015;11:e1005242. [Crossref] [PubMed]
- Arora S, Ranade AR, Tran NL, et al. MicroRNA-328 is associated with (non-small) cell lung cancer (NSCLC) brain metastasis and mediates NSCLC migration. Int J Cancer 2011;129:2621-31. [Crossref] [PubMed]
- Smeltzer MP, Lee YS, Faris M, Div NR, et al. Trends in Accuracy and Comprehensiveness of Pathology Reports for Resected NSCLC in a High Mortality Area of the United States. J Thorac Oncol 2021;16:1663-71. [Crossref] [PubMed]
- Borzi C, Ganzinelli M, Caiola E, et al. LKB1 Down-Modulation by miR-17 Identifies Patients With NSCLC Having Worse Prognosis Eligible for Energy-Stress-Based Treatments. J Thorac Oncol 2021;16:1298-311. [Crossref] [PubMed]
- Nishio M, Barlesi F, West H, et al. Atezolizumab Plus Chemotherapy for First-Line Treatment of Nonsquamous NSCLC: Results From the Randomized Phase 3 IMpower132 Trial. J Thorac Oncol 2021;16:653-64. [Crossref] [PubMed]
- Zhang B, Pan X, Cobb GP, et al. microRNAs as oncogenes and tumor suppressors. Dev Biol 2007;302:1-12. [Crossref] [PubMed]
- Yu L, Sui B, Fan W, et al. Exosomes derived from osteogenic tumor activate osteoclast differentiation and concurrently inhibit osteogenesis by transferring COL1A1-targeting miRNA-92a-1-5p. J Extracell Vesicles 2021;10:e12056. [Crossref] [PubMed]
- Zhang YH, Jin M, Li J, et al. Identifying circulating miRNA biomarkers for early diagnosis and monitoring of lung cancer. Biochim Biophys Acta Mol Basis Dis 2020;1866:165847. [Crossref] [PubMed]
- Zhang J, Lou W. A Key mRNA-miRNA-lncRNA Competing Endogenous RNA Triple Sub-network Linked to Diagnosis and Prognosis of Hepatocellular Carcinoma. Front Oncol 2020;10:340. [Crossref] [PubMed]
- Yin H, Wang Y, Wu Y, et al. EZH2-mediated Epigenetic Silencing of miR-29/miR-30 targets LOXL4 and contributes to Tumorigenesis, Metastasis, and Immune Microenvironment Remodeling in Breast Cancer. Theranostics 2020;10:8494-512. [Crossref] [PubMed]
- Sharma P, Dando I, Strippoli R, et al. Nanomaterials for Autophagy-Related miRNA-34a Delivery in Cancer Treatment. Front Pharmacol 2020;11:1141. [Crossref] [PubMed]
- Fehlmann T, Kahraman M, Ludwig N, et al. Evaluating the Use of Circulating MicroRNA Profiles for Lung Cancer Detection in Symptomatic Patients. JAMA Oncol 2020;6:714-23. [Crossref] [PubMed]
- Jiang N, Zou C, Zhu Y, et al. HIF-1ɑ-regulated miR-1275 maintains stem cell-like phenotypes and promotes the progression of LUAD by simultaneously activating Wnt/β-catenin and Notch signaling. Theranostics 2020;10:2553-70. [Crossref] [PubMed]
- Zhong S, Golpon H, Zardo P, et al. miRNAs in lung cancer. A systematic review identifies predictive and prognostic miRNA candidates for precision medicine in lung cancer. Transl Res 2021;230:164-96. [Crossref] [PubMed]
- Daugaard I, Sanders KJ, Idica A, et al. miR-151a induces partial EMT by regulating E-cadherin in NSCLC cells. Oncogenesis 2017;6:e366. [Crossref] [PubMed]
- Nikolic I, Elsworth B, Dodson E, et al. Discovering cancer vulnerabilities using high-throughput micro-RNA screening. Nucleic Acids Res 2017;45:12657-70. [Crossref] [PubMed]
- Rodriguez-Pascual F, Rosell-Garcia T. Lysyl Oxidases: Functions and Disorders. J Glaucoma 2018;27:S15-9. [Crossref] [PubMed]
- Lucero HA, Kagan HM. Lysyl oxidase: an oxidative enzyme and effector of cell function. Cell Mol Life Sci 2006;63:2304-16. [Crossref] [PubMed]
- Li R, Wang Y, Zhang X, et al. Exosome-mediated secretion of LOXL4 promotes hepatocellular carcinoma cell invasion and metastasis. Mol Cancer 2019;18:18. [Crossref] [PubMed]
- Zhang Y, Jiang WL, Yang JY, et al. Downregulation of lysyl oxidase-like 4 LOXL4 by miR-135a-5p promotes lung cancer progression in vitro and in vivo. J Cell Physiol 2019;234:18679-87. [Crossref] [PubMed]
- Li RK, Zhao WY, Fang F, et al. Lysyl oxidase-like 4 (LOXL4) promotes proliferation and metastasis of gastric cancer via FAK/Src pathway. J Cancer Res Clin Oncol 2015;141:269-81. [Crossref] [PubMed]
- Wu G, Guo Z, Chang X, et al. LOXL1 and LOXL4 are epigenetically silenced and can inhibit ras/extracellular signal-regulated kinase signaling pathway in human bladder cancer. Cancer Res 2007;67:4123-9. [Crossref] [PubMed]


