
B3GNT3 as a prognostic biomarker and correlation with immune cell infiltration in lung adenocarcinoma
Introduction
Lung cancer is the most prevalent cancer with high mortality, accounting for more than 18 million new cancer cases and 9 million cancer-related deaths worldwide (1). Lung adenocarcinoma (LUAD) is the primary subtype of lung cancer, accounting for approximately 50% of all lung cancer patients (2). Over the past decade, integrated treatments such as surgical resection, chemotherapy, radiotherapy, and molecular-targeted therapy have been provided for LUAD patients (3-5), but overall survival (OS) rates have not significantly improved. Targeted therapies using tyrosine kinase inhibitors (TKI) such as gefitinib, erlotinib, and crizotinib have revolutionized the treatment of the adenocarcinoma subtype (6-9). However, efficacy is limited by acquisition of mutations, resulting in cancer progression for the majority of LUAD patients. It is vital to understand the mechanisms underlying LUAD progression and explore potential biomarkers.
B3GNT3, Beta-1,3-N-acetylglucosaminyltransferase 3, is a type II transmembrane protein located in the Golgi apparatus and is essential for forming extended core 1 oligosaccharides (10,11). Numerous studies have reported the involvement of B3GNT3 in malignant transformation and B3GNT3 expression is strongly correlated with PD-L1 levels and EGFR mutation status (12-19). It has been indicated that B3GNT3 expression is strongly correlated with PD-L1 levels and EGFR mutation status. High expression of B3GNT3 is associated with advanced histologic grade and poorer OS. However, in neuroblastoma B3GNT3 expression inhibits cell migration and invasion and predicts favorable outcomes (14). It has been determined that the tumor-infiltrating immune cells are correlated with the development, progression and prognosis of LUAD (20). Moreover, DNA methylation is associated with the expression of gene and reduces tumor immunity and the prognosis of patients with LUAD (21). However, the methylation and B3GNT3 remains unclear. Therefore, the role of B3GNT3 in LUAD is controversial.
Our study detected that B3GNT3 was more highly expressed in tumor tissues than in normal tissues and more in LUAD cell lines than in human bronchial epithelial cells. B3GNT3 was also related to poorer OS for LUAD patients. Furthermore, B3GNT3 was identified as an independent prognostic factor. Strikingly, B3GNT3 was involved in immune cell infiltration and might correlate with VTCN1, VSIR, CTLA-4, and CD276 for LUAD immunotherapy. Furthermore, the results suggested a strong correlation of B3GNT3 DNA methylation with mRNA levels and survival. Therefore, we identified B3GNT3 as an important biomarker and validated the prognostic and predictive values for LUAD. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-493/rc).
Methods
Data collection and bioinformatic analysis
The RNA expression profile and corresponding clinical characteristics were downloaded from The Cancer Genome Atlas (TCGA). GSE68465 (22) from the GEO database had 442 lung adenocarcinomas enrolled for mRNA. To detect the protein levels of B3GNT3, the Human Protein Atlas (HPA) database was accessed for the immunohistochemistry results for tumors and normal tissues. To analyze genomic alternations, we measured the alteration rates based on the cBioPortal tool. Furthermore, we examined the association between B3GNT3 expression and DNA methylation using the MEXPRESS tool (http://mexpress.be/) (23). Data on gene methylation were downloaded from the UCSC Xena browser (https://xena.ucsc.edu/). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture
LUAD cell lines (A549 and PC9) and human bronchial epithelial cell line BEAS-2B were purchased from the American Type Culture Collection. Cells were maintained in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum (Gibco, CA, USA) at 37 ℃ in a humidified incubator with 5% CO2 at 37 ℃.
RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells by TRIzol reagent (Invitrogen, NY, USA). cDNAs were generated from RNA via reverse transcription using PrimeScript RT Master Mix (TIANGEN, Beijing, China). The primers for GAPDH were as follows: Forward 5'-GTCTCCTCTGAC TTCAACAGCG-3' and Reverse 5'-ACCACCCTGTTG CTGTAGCCAA-3'. The primers for B3GNT3 were Forward 5'-TATGTGTCTGGAGCTTGAGG-3', Reverse 5'-AAGGATGTGTAGGAGTTCGC-3'. qRT-PCR was performed using the SYBR Premix Ex Taq reaction system according to the manufacturer’s instructions. The levels of mRNA were measure by calculating 2−ΔΔCT.
Western blot assay
Total proteins were extracted from cells, and the same amounts were loaded on the 12% SDS-PAGE gels and then blotted onto polyvinylidene fluoride membranes. After blocking with non-fat milk for 1.5 h, the blots were incubated with the primary antibodies for B3GNT3 (NBP1-32539; Novus Biologicals, USA) and GAPDH (ab8245, Abcam, UK) at 4 ℃ overnight. Subsequently, the membranes were incubated with secondary antibodies for 1 h.
Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses and protein-protein interaction (PPI)
Differentially expressed genes (DEGs) were identified by the criteria of adjusted P value <0.05 and log 2-fold change (FC) >0.5. We performed GO enrichment analysis comprising biological process (BP), cellular components (CC), and molecular function (MF), and KEGG for DEGs to explore the utilities of the biological functions using the R package “clusterProfiler” (24,25). Gene expression profiles of LUAD patients were divided into high and low groups according to the median value of expression of B3GNT3. Gene set enrichment analysis (GSEA) was utilized to detect possible mechanisms underlying the effect of B3GNT3 expression on LUAD prognosis. In addition, gene set variation analysis (GSVA) was used to detect the pathways most associated with B3GNT3. A PPI network helped us explore novel molecules with B3GNT3. The STRING (https://string-db.org/) database is a tool for searching the interactions among different proteins. Significantly associated genes with weight value >0.8 were collected to construct the PPI network.
Identification of an independent factor for LUAD patients
Univariate and multivariate regression analyses, and a meta-analysis using the TCGA and GSE68465 for the hazard ratio (HR) in LUAD were performed to determine an independent prognostic factor.
Correlation between gene expression and immune cell infiltration
The tool CIBERSORT (Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts) was applied to investigate the correlation between B3GNT3 and 22 immune cells. We assessed the correlation between expression of B3GNT3, copy number alteration (CNA), and tumor-infiltrating immune cells by TIMER (https://cistrome.shinyapps.io/timer/), which included different types of cancer samples accessible in the TCGA. We examined the association between B3GNT3 and key checkpoints for its possible use in immune therapy.
Construction and evaluation of a predictive nomogram
All independent prognostic parameters and relevant clinical parameters were included in the construction of a prognostic nomogram via a stepwise Cox regression model to predict 1-, 3-, and 5-year overall survival of patients in the TCGA dataset. A nomogram was established via the RMS package in R (26-28).
Cell transfection
RNA interference of B3GNT3 was performed using small interfering RNA (siRNA). Cells were transfected with non-specific scramble (scr) siRNA as a control and siB3GNT3 using Lipofectamine 3000 (Invitrogen, CA, USA). The siRNAs used were: siB3GNT3 sense 5'-CAGACAACAUGGUCUUCUAtt-3' and antisense 5'-TAGAAGACCATGTTGTCTGtg-3'.
Cell proliferation, cloning, and invasion
Proliferative ability was determined using 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay according to the manufacturer’s instructions. A microplate reader was used to examine the absorbance values at 490 nm. One thousand cells per well were seeded in 6-well plates and cultured for 2 weeks. Colonies were immobilized with 10% formaldehyde for 5 min and then stained with 1.0% crystal violet for 30 s. For the invasion assay, after 48 h of incubation, cells in the lower chamber were stained with 0.1% crystal violet and counted under a microscope with 6 different fields per filter.
Statistical analysis
All the values are presented as mean ± standard deviation (SD). A t-test and one-way analysis of variance were applied to assess the differences between groups. P<0.05 was considered a significant difference.
Results
Levels of B3GNT3 mRNA in LUAD
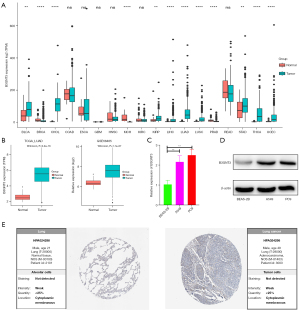
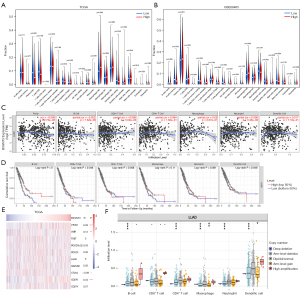
Gene expression analyses using the UCSC Xena database indicated that B3GNT3 expression was significantly higher in bladder urothelial carcinoma (BLCA), cholangiocarcinoma (CHOL), kidney renal papillary (KIRP), liver hepatocellular carcinoma (LIHC), LUAD, lung squamous cell carcinoma (LUSC), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC), but the levels of B3GNT3 mRNA were much lower in breast invasive carcinoma (BRCA), kidney chromophobe (KICH), and prostate adenocarcinoma (PRAD), compared with corresponding normal tissues (Figure 1A). Moreover, based on both the TCGA-LUAD database and GSE68465 dataset, B3GNT3 was overexpressed in LUAD tissues (Figure 1B). In addition, the expression of B3GNT3 was much higher in A549 and PC9 cells than in BEAS-2B cells (Figure 1C). Moreover, B3GNT3 protein was lower in BEAS-2B cells than in A549 and PC9 cells (Figure 1D). Furthermore, the HPA determined that B3GNT3 protein levels in LUAD tissues were much higher than in normal lung tissues (Figure 1E). The B3GNT3 protein might be distributed in the cytoplasm and membranes. Thus, all data indicated high expression of B3GNT3 might be associated with LUAD.
Prognostic value of B3GNT3 in LUAD
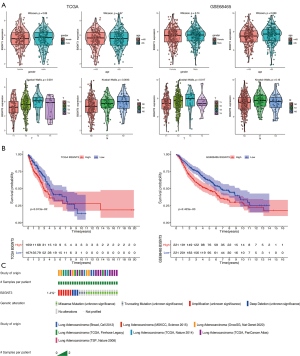
The characteristics of LUAD patients with B3GNT3 levels from the TCGA are presented in Table 1. In the TCGA-LUAD database, among 4 clinicopathological features, high expression of B3GNT3 was associated with pathological T and pathological N categories, but not with age or sex. Moreover, a consistent finding was determined from the GSE68465 dataset (Figure 2A). To further explore the prognosis of B43GNT3, a total of 332 LUAD patients from the TCGA database were divided into two groups according to the expression of B3GNT3. The Kaplan-Meier curve suggested that patients with high expression suffered worse OS rates than patients with low expression, which was similar to the results from GSE68465 (Figure 2B). To detect the mutation of B3GNT3, the rates of genetic alteration were 1.4% in patients from 7 different studies (Figure 2C). Therefore, the evidence suggested high levels of B3GNT3 are related to poor prognosis.
Table 1
| Patients | High expression (N=118) | Low expression (N=119) | P value |
|---|---|---|---|
| Sex, n (%) | 0.842 | ||
| Female | 54 (45.8) | 57 (47.9) | |
| Male | 64 (54.2) | 62 (52.1) | |
| Age (years), n (%) | 0.851 | ||
| <65 | 52 (44.1) | 50 (42.0) | |
| >65 | 66 (55.9) | 69 (58.0) | |
| Stage, n (%) | 0.00966 | ||
| I | 48 (40.7) | 68 (57.1) | |
| II | 27 (22.9) | 30 (25.2) | |
| III | 30 (25.4) | 16 (13.4) | |
| IV | 13 (11.0) | 5 (4.2) | |
| T, n (%) | 0.258 | ||
| T1 | 32 (27.1) | 39 (32.8) | |
| 2 | 65 (55.1) | 68 (57.1) | |
| 3 | 14 (11.9) | 6 (5.0) | |
| 4 | 7 (5.9) | 6 (5.0) | |
| N, n (%) | 0.124 | ||
| 0 | 64 (54.2) | 81 (68.1) | |
| 1 | 29 (24.6) | 23 (19.3) | |
| 2 | 24 (20.3) | 15 (12.6) | |
| 3 | 1 (0.8) | 0 (0.0) | |
| M, n (%) | 0.215 | ||
| 0 | 105 (89.0) | 114 (95.8) | |
| 1 | 10 (8.5) | 3 (2.5) | |
| 1a | 1 (0.8) | 1 (0.8) | |
| 1b | 2 (1.7) | 1 (0.8) | |
| Cigarettes per day | 0.624 | ||
| Mean (SD) | 2.38 (1.52) | 2.28 (1.50) | |
| Median (min, max) | 2.19 (0.00822, 8.44) | 2.05 (0.110, 8.05) | |
| Status, n (%) | 0.935 | ||
| Alive | 73 (61.9) | 72 (60.5) | |
| Dead | 45 (38.1) | 47 (39.5) |
T, tumor; N, node; M, metastasis; SD, standard deviation.
Functional enrichment of DEGs based on B3GNT3
To identify the DEGs based on the expression of B3GNT3, DEGs were obtained by RNA-seq from the GSE68465 dataset. A total of 165 DEGs (106 upregulated and 59 downregulated genes) were identified (Figure 3A, https://cdn.amegroups.cn/static/public/atm-22-493-1.xls). The top 100 DEGs are shown in Figure 3B. To detect the functional enrichment of the DEGs, we used GO annotation, including BP, CC, and MF. Enriched BPs were O-glycan processing, epidermis development and protein O-linked glycosylation. In terms of CC, the DEGs were enriched with laminin complex, Golgi lumen, and intermediate filament, and for MF, they were enriched in the structural constituents of the cytoskeleton, aromatase activity and serine-type endopeptidase inhibitor activity. KEGG analysis indicated that the DEGs were most enriched in the interleukin-17 signaling pathway, amoebiasis, and retinol metabolism (Figure 3C). Furthermore, GSEA demonstrated that the DEGs were associated with cell cycle, P53 signaling pathway, and valine leucine and isoleucine degradation (Figure 3D). In addition, GSVA suggested that these DEGs highly affected the development of LUAD (Figure 3E). Our data revealed that DEGs could be involved in the development of LUAD.

Identification of key candidate genes
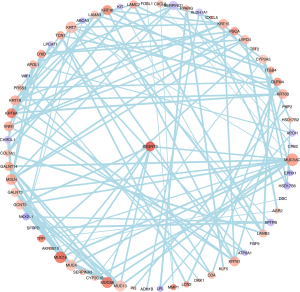
A protein-protein interaction was constructed to analyze the possible targets of B3GNT. In total, 67 genes were identified as key genes that were highly associated with B3GNT3 (Figure 4). The genes that were significantly correlated with B3GNT3 were GJB3, ITGB4, PTPRH, KRT19 and LAMC2.
An independent factor for the prognosis of LUAD
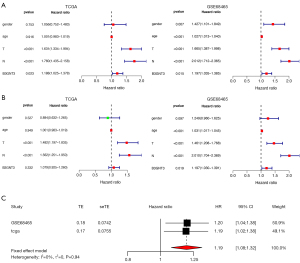
To determine whether B3GNT3 was an independent factor, univariate Cox analysis was performed. Our data showed that pathological T and N categories, and B3GNT3 were significantly associated with survival in the TCGA. In the GSE68465 cohort, sex, pathological T and N categories, and B3GNT3 were highly related to LUAD survival rates (Figure 5A). However, the multivariate Cox regression model indicated that T and N categories were significantly associated with prognosis. T and N categories, and B3GNT3 highly correlated with the survival of LUAD patients (Figure 5B). Additionally, from our meta-analysis the HR was 1.19 (confidence interval: 1.08–1.32, I2=0) (Figure 5C). Therefore, B3GNT3 was likely to be an independent prognosticator for LUAD.
Correlation between the expression of B3GNT3 and immune cell infiltration and checkpoints
To further determine the role of B3GNT3 in the tumor microenvironment (TME), we explored the association between B3GNT3 and 22 immune cells. Notably, in the TCGA database, plasma cells and resting CD4 memory T cells were enriched in the samples of the low-B3GNT3 group, whereas M0 macrophages were enriched in the samples of the high-B3GNT3 group (Figure 6A). In the GSE68465 dataset, a significant association was found between the samples in the low-B3GNT3 group and gamma-delta T cells and resting dendritic cells. The samples in the high-B3GNT3 group significantly correlated with resting natural killer cells, M0 macrophages, activated dendritic cells, and neutrophils (Figure 6B). Additionally, our findings indicated that B3GNT3 were associated with tumor purity, B cells, and neutrophils (Figure 6C). B cells and dendritic cells were associated with survival of LUAD (Figure 6D). We also determined the association of B3GNT3 and immune checkpoints by referring to the TCGA database. The levels of B3GNT3 might correlate with VTCN1 (B7-H4), VSIR (B7-H5), CTLA-4, and CD276 (B7-H3) (Figure 6E). Furthermore, we explored the effect of somatic CNA on the level of infiltration of different immune cells. The arm-level deletion of B3GNT3 CNA was significantly associated with B cells, CD4+ cells, macrophages, neutrophils, and dendritic cells. Moreover, arm-level deletion of B3GNT3 CNA greatly affected macrophages and dendritic cells (Figure 6F). Therefore, our results suggested that B3GNT3 correlated with immune cell infiltration and immune checkpoints.
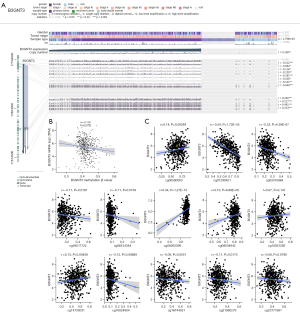
Association between DNA methylation of B3GNT3 and prognosis of LUAD
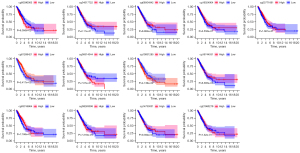
The association between B3GNT3 expression and DNA methylation was shown in Figure 7A. The association between DNA methylation and the expression of mRNA in LUAD was analyzed (r=–0.116, P=0.0133), indicating that B3GNT3 expression negatively correlated with the DNA methylation level (Figure 7B). A significant association between 10 methylation sites and the level of B3GNT3 mRNA was found (Figure 7C). Furthermore, two CpG island methylation levels (cg01289421 and cg10831285) of B3GNT3 significantly influenced the OS rate of LUAD patients (Figure 8).
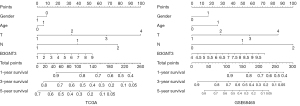
Nomogram construction
To identify an individual’s risk, a nomogram was created based on the TCGA and GSE68465 dataset to predict the 1-, 3-, and 5-year survival rates. The nomogram contained 7 prognostic factors, including sex, age, pathological T and N categories B3GNT3 (Figure 9). After a comprehensive assessment of prognostic value, the risk model was considered as a biomarker, exerting predictive value for LUAD patients
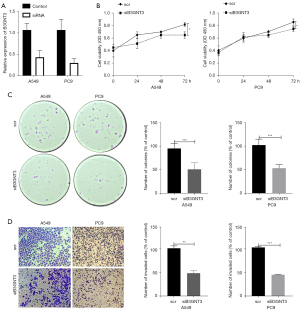
Effect of knockdown of B3GNT3 on LUAD cell proliferation, cloning, and invasive ability
To understand the functions of B3GNT3, we transfected siRNA into LUAD cells (A549 and PC9) to knockdown the mRNA levels of B34GNT3. It was showed that the mRNA of B3GNT3 was highly downregulated by as much as 67% in A549 and 70% in PC9 (Figure 10A). MTT assay measured cell viability after suppression of B3GNT3, revealing a significant downregulation of cell viability of A549 and PC9 cells after 72 h (Figure 10B). Meanwhile, we found the number of cell colony formation units was dramatically reduced after downregulation of B3GNT3 in A549 and PC9 cells (Figure 10C). Our transwell assays revealed that B3GNT3 knockdown significantly suppressed cell invasion in A549 and PC9 cells (Figure 10D). Therefore, LUAD cell proliferation, cloning, and invasion were significantly inhibited after downregulation of B3GNT3.
Discussion
Lung cancer led to more than 140,000 cancer-related deaths in the USA in 2019 (2). The emerging targeted therapy has provided the possibility of prolonging survival, but more novel therapeutic targets urgently need to be identified. B3GNT3 has been reported as an oncogene in a few cancer cases and is involved in cell proliferation and lymphocyte trafficking (12,16,18). However, there is no integrated and comprehensive study in this area, particularly regarding the methylation of B3GNT3 in LUAD patients.
In our current work, the mRNA levels of B3GNT3 were much higher in tumor tissues than in adjacent normal tissues from three different databases, which was also validated in our cell lines. The levels of B3GNT3 protein were determined using the HPA database and western blot, indicating higher expression in LUAD tissues and LUAD cells than in normal lung tissues and human bronchial epithelial cells, respectively. Consistent with our findings, B3GNT3 was revealed as a carcinogenic factor by presenting higher expression in pancreatic cancer (16), early-stage cervical cancer (15), neuroblastoma (14), and endometrial cancer tissues than in normal tissues (29). In our study, high levels of B3GNT3 were associated with poor prognosis with advanced stage and higher mortality rates, which was also indicated in previous lung cancer (19) and non-small cell lung cancer studies (12). Moreover, the genetic alteration of B3GNT3 was 1.4% in LUAD patients from several datasets, which suggests the possibility of improving the prognosis by targeting B3GNT3. To determine the possible mechanisms of B3GNT3, we conducted GESA and GSVA to analyze DEGs, which were enriched in cancer development, emphasizing the carcinogenic role of B3GNT3 and supporting our conclusions. Based on the results of our univariate analysis, multivariate Cox regression analysis, and meta-analysis from the TCGA and GSE68465, we concluded that B3GNT3 was an independent factor of the prognosis of LUAD, similar to its role in non-small cell lung cancer and early-stage cervical cancer (15). We consider that B3GNT3 was validated as an independent carcinogenic factor that significantly affects the prognosis of LUAD patients.
Increasing attention is being given to the TME, including immune cell infiltration. Our study found that M0 macrophages tend to be enriched in patients with high expression of B3GNT3 in both the TCGA-LUAD and GEO databases. Abundance of M0 macrophages has been linked to significantly poor OS rates for LUAD patients in other studies (30,31). B3GNT3 also correlated with B cells and neutrophils, which were significantly associated with LUAD survival based on our data. Our conclusions were similar to one previous study asserting that tumor-infiltrating B lymphocytes result in favorable outcomes (32). The potential mechanisms need to be investigated by subsequent fundamental research focusing on the functions of B cells in LUAD. Consistent with our research, another study demonstrated that neutrophils alter angiogenesis and increase hypoxia to maintain a deleterious TME, which indicates a novel role of neutrophils (33). Furthermore, dendritic cell paucity leads to dysfunctional immune surveillance in pancreatic cancer, playing an important role in tumor immunity (34). Based on these other studies, we believe that B3GNT3 might affect M0 macrophages, B cells, neutrophils, and dendritic cells, leading to a poor prognosis for LUAD.
We also explored the association between B3GNT3 and several checkpoints, including VTCN1 (B7-H4), VSIR (B7-H5), TIGIT, PDCD1LG2 (PD-L2), PDCD1 (PD-1), LAG3, HAVCR2 (TIM-3), CTLA4, CD276 (B7-H3), and CD274 (PD-L1). Our results demonstrated that B3GNT3 highly correlated with VTCN1 (B7-H4) and CD276 (B7-H3). B7-H3 and B7-H4 were validated as inhibiting the activation and function of T cells, potently suppressing the proliferation, cytokine production, and cytotoxicity of activated T cells (35,36). Enoblituzumab, a monoclonal antibody targeting B7-H3, was the first agent to enter clinical practice, and has shown acceptable tolerability and efficacious results in combination with pembrolizumab in patients with various solid malignancies (37). Thus, the data suggest that B3GNT3 might correlate with multiple biomarkers for immunotherapy in LUAD, although further validation involving larger clinical cohorts is urgently needed.
To our knowledge, this study is the first to focus on the methylation of B3GNT3. DNA methylation is a common modification that participates in the initiation and process of LUAD. Our study found a significant association of B3GNT3 DNA methylation with mRNA expression at 10 of 13 analyzed CpG sites, and with survival at 2 of 13 sites for LUAD patients. In addition, our nomogram indicated that B3GNT3 was a predictive biomarker in LUAD. Similarly, TNFRSF9 DNA methylation acts as a novel biomarker, regulating mRNA expression (38). In the early-stage breast cancer, S100P methylation in the blood was validated to have predictive value. To validate the role of B3GNT3 in LUAD, we performed cell proliferation, cloning, and invasion studies, which supported our hypothesis. One possible mechanism is that B3GNT3 binds to miR-149-5p directly to promote the development of LUAD (19). Thus, B3GNT3 could be a prognostic and predictive biomarker for LUAD.
Conclusions
High expression of B3GNT3 was related to poor prognosis, acting as an independent factor for LUAD, and involved in both immune cell infiltration and several immune checkpoints. Moreover, a significant correlation of B3GNT3 DNA methylation with mRNA expression and survival was determined, showing predictive value for LUAD. Further research is needed to explore the potential mechanisms and therapeutic value of B3GNT3 in LUAD.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-493/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-493/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-493/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Chen D, Wang R, Yu C, et al. FOX-A1 contributes to acquisition of chemoresistance in human lung adenocarcinoma via transactivation of SOX5. EBioMedicine 2019;44:150-61. [Crossref] [PubMed]
- Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol 2011;12:175-80. [Crossref] [PubMed]
- Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 2016;66:271-89. [Crossref] [PubMed]
- Cai W, Lin D, Wu C, et al. Intratumoral Heterogeneity of ALK-Rearranged and ALK/EGFR Coaltered Lung Adenocarcinoma. J Clin Oncol 2015;33:3701-9. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Turke AB, Zejnullahu K, Wu YL, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell 2010;17:77-88. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Yeh JC, Hiraoka N, Petryniak B, et al. Novel sulfated lymphocyte homing receptors and their control by a Core1 extension beta 1,3-N-acetylglucosaminyltransferase. Cell 2001;105:957-69. [Crossref] [PubMed]
- Mitoma J, Petryniak B, Hiraoka N, et al. Extended core 1 and core 2 branched O-glycans differentially modulate sialyl Lewis X-type L-selectin ligand activity. J Biol Chem 2003;278:9953-61. [Crossref] [PubMed]
- Leng X, Wei S, Mei J, et al. Identifying the prognostic significance of B3GNT3 with PD-L1 expression in lung adenocarcinoma. Transl Lung Cancer Res 2021;10:965-80. [Crossref] [PubMed]
- Gao L, Zhang H, Zhang B, et al. B3GNT3 overexpression is associated with unfavourable survival in non-small cell lung cancer. J Clin Pathol 2018;71:642-7. [Crossref] [PubMed]
- Kong K, Zhao Y, Xia L, et al. B3GNT3: A prognostic biomarker associated with immune cell infiltration in pancreatic adenocarcinoma. Oncol Lett 2021;21:159. [Crossref] [PubMed]
- Ho WL, Che MI, Chou CH, et al. B3GNT3 expression suppresses cell migration and invasion and predicts favorable outcomes in neuroblastoma. Cancer Sci 2013;104:1600-8. [Crossref] [PubMed]
- Zhang W, Hou T, Niu C, et al. B3GNT3 Expression Is a Novel Marker Correlated with Pelvic Lymph Node Metastasis and Poor Clinical Outcome in Early-Stage Cervical Cancer. PLoS One 2015;10:e0144360. [Crossref] [PubMed]
- Barkeer S, Chugh S, Karmakar S, et al. Novel role of O-glycosyltransferases GALNT3 and B3GNT3 in the self-renewal of pancreatic cancer stem cells. BMC Cancer 2018;18:1157. [Crossref] [PubMed]
- Zhuang H, Zhou Z, Zhang Z, et al. B3GNT3 overexpression promotes tumor progression and inhibits infiltration of CD8+ T cells in pancreatic cancer. Aging (Albany NY) 2020;13:2310-29. [Crossref] [PubMed]
- Li CW, Lim SO, Chung EM, et al. Eradication of Triple-Negative Breast Cancer Cells by Targeting Glycosylated PD-L1. Cancer Cell 2018;33:187-201.e10. [Crossref] [PubMed]
- Sun Y, Liu T, Xian L, et al. B3GNT3, a Direct Target of miR-149-5p, Promotes Lung Cancer Development and Indicates Poor Prognosis of Lung Cancer. Cancer Manag Res 2020;12:2381-91. [Crossref] [PubMed]
- Director's Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med 2008;14:822-7. [Crossref] [PubMed]
- Koch A, De Meyer T, Jeschke J, et al. MEXPRESS: visualizing expression, DNA methylation and clinical TCGA data. BMC Genomics 2015;16:636. [Crossref] [PubMed]
- Harris MA, Clark J, Ireland A, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 2004;32:D258-61. [Crossref] [PubMed]
- Kanehisa M, Sato Y, Kawashima M, et al. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 2016;44:D457-62. [Crossref] [PubMed]
- Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol 2008;26:1364-70. [Crossref] [PubMed]
- Jeong SH, Kim RB, Park SY, et al. Nomogram for predicting gastric cancer recurrence using biomarker gene expression. Eur J Surg Oncol 2020;46:195-201. [Crossref] [PubMed]
- Zhong H, Wang J, Zhu Y, et al. Comprehensive Analysis of a Nine-Gene Signature Related to Tumor Microenvironment in Lung Adenocarcinoma. Front Cell Dev Biol 2021;9:700607. [Crossref] [PubMed]
- Wang JS, Ruan F, Guo LZ, et al. B3GNT3 acts as a carcinogenic factor in endometrial cancer via facilitating cell growth, invasion and migration through regulating RhoA/RAC1 pathway-associated markers. Genes Genomics 2021;43:447-57. [Crossref] [PubMed]
- Zhang L, Chen J, Cheng T, et al. Identification of the key genes and characterizations of Tumor Immune Microenvironment in Lung Adenocarcinoma (LUAD) and Lung Squamous Cell Carcinoma (LUSC). J Cancer 2020;11:4965-79. [Crossref] [PubMed]
- Mo Z, Yu L, Cao Z, et al. Identification of a Hypoxia-Associated Signature for Lung Adenocarcinoma. Front Genet 2020;11:647. [Crossref] [PubMed]
- Wang SS, Liu W, Ly D, et al. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cell Mol Immunol 2019;16:6-18. [Crossref] [PubMed]
- Faget J, Groeneveld S, Boivin G, et al. Neutrophils and Snail Orchestrate the Establishment of a Pro-tumor Microenvironment in Lung Cancer. Cell Rep 2017;21:3190-204. [Crossref] [PubMed]
- Hegde S, Krisnawan VE, Herzog BH, et al. Dendritic Cell Paucity Leads to Dysfunctional Immune Surveillance in Pancreatic Cancer. Cancer Cell 2020;37:289-307.e9. [Crossref] [PubMed]
- Sica GL, Choi IH, Zhu G, et al. B7-H4, a molecule of the B7 family, negatively regulates T cell immunity. Immunity 2003;18:849-61. [Crossref] [PubMed]
- Chapoval AI, Ni J, Lau JS, et al. B7-H3: a costimulatory molecule for T cell activation and IFN-gamma production. Nat Immunol 2001;2:269-74. [Crossref] [PubMed]
- Loo D, Alderson RF, Chen FZ, et al. Development of an Fc-enhanced anti-B7-H3 monoclonal antibody with potent antitumor activity. Clin Cancer Res 2012;18:3834-45. [Crossref] [PubMed]
- Fröhlich A, Loick S, Bawden EG, et al. Comprehensive analysis of tumor necrosis factor receptor TNFRSF9 (4-1BB) DNA methylation with regard to molecular and clinicopathological features, immune infiltrates, and response prediction to immunotherapy in melanoma. EBioMedicine 2020;52:102647. [Crossref] [PubMed]
- Yang R, Stöcker S, Schott S, et al. The association between breast cancer and S100P methylation in peripheral blood by multicenter case-control studies. Carcinogenesis 2017;38:312-20. [Crossref] [PubMed]
(English Language Editor: K. Brown)


