
GeneChip expression profiling identified OLFML2A as a potential therapeutic target in TNBC cells
Introduction
Olfactomedin-like-2A (OLFML2A), also known as photomedin-1, is a glycoprotein belonging to subfamily IV of the olfactomedin family (1). The human OLFML2A gene, located on chromosome 9q33.3, comprises at least eight exons and spans 37.7 kb. The gene product of OLFML2A is located in the extracellular region and is involved in extracellular matrix (ECM) organization (2). Among the ECM components, the OLFML2A protein preferentially binds to chondroitin sulfate-E and heparin (3). In humans, OLFML2A is expressed in the breast, bronchus, caudate, cerebral cortex, colon, duodenum, eye, gallbladder, heart muscle, hippocampus, kidney, lung, pancreas, rectum, seminal vesicle, skeletal muscle, skin, small intestine, stomach, and testis under physiological conditions according to data from The Human Protein Atlas (4). OLFML2A was first described in mice in 2005 (3). In the mouse retina, OLFML2A is predominantly expressed in the outer segment of photoreceptor cells. Its mRNA has been detected in the retina, cornea and lens of the eye, and its protein is expressed in ganglion cells and retina cells in baboons and humans (5). It is also highly expressed in human podocytes, which contribute to the formation of the glomerular crescent (6). The OLFML2A gene is also upregulated in the skin of premenopausal women compared with that in postmenopausal women (7). Its expression is higher in the annulus fibrosus than in nucleus pulposus cells in the mature human intervertebral disc, and it serves as a marker of the annulus fibrosus (8). OLFML2A gene expression is also upregulated in the osteogenic differentiation of adipose-derived stem cells in humans (9). Interestingly, cancer cells (H460 and U87MG) can promote OLFML2A gene expression in human adipose tissue-derived mesenchymal stem cells (10). Angiotensin II also decreases OLFML2A mRNA expression in rat cardiac fibroblasts (11). In zebrafish, olfactomedin 2 encodes a secreted glycoprotein that is involved in nervous system development, and the downregulation of this gene adversely affects the development of the optic tectum and eyes (12,13). Gene ontology (GO) annotations revealed that the OLFML2A gene plays a key role in protein homodimerization activity and ECM binding.
However, OLFML2A gene expression is intricate under pathological conditions. OLFML2A protein can be detected in human neoplastic tissues, such as breast, cervical, colorectal, liver, lung, ovarian, pancreatic, and stomach cancers (14). Its expression is increased in renal cell carcinoma compared to that in normal renal tissue (15). OLFML2A also endows tumor cells with higher migration activity in renal cell carcinoma ACHN, Caki1, Caki2, and KIJ265T cell lines (15). In addition, the OLFML2A gene is upregulated in three-dimensional (3D) human bladder T24 cells in the host microenvironment compared to that in two-dimensional (2D) cultures (16). Its expression is also upregulated in chemotherapy-resistant alveolar soft part sarcoma compared to that in normal sarcoma (17). It is also upregulated in mouse thyroid side population cells, which possess stem cell characteristics, compared to in non-side population cells (18). It seems that OLFML2A expression is correlated with the malignant degree of these cancers. The expression of the OLFML2A gene is lower in a highly metastatic subclone of adenoid cystic carcinoma compared to that in its parental subclone (19). It is also downregulated in spheroid cells compared to monolayer cells in cervical HeLa cells (20). The expression of its mRNA is higher in endometrial carcinoma compared to that in the normal endometrium (21). In addition, its expression is increased in EGFR-mutant non-small cell lung cancer HCC827 cells treated with erlotinib compared to DMSO-treated controls (22). Kaplan-Meier plots have shown that OLFML2A expression is unfavorable in some cancers, such as stomach, testis, urothelial, melanoma, glioma, and breast cancers (14). However, it is favorable for patients with pancreatic and cervical cancers (14). In particular, OLFML2A is prognostic, and the elevated expression of OLFML2A is unfavorable in renal cancer patients, with a P value of 0.000037 (14). Therefore, this evidence shows that the OLFML2A gene is heterogenous.
Recently, Peng (23) found that the expression of OLFML2A mRNA was higher in hepatic carcinoma than in normal liver tissue, and the knockdown of OLFML2A inhibited proliferation and promoted apoptosis in hepatic carcinoma cells. In addition, a short hairpin RNA (shRNA) targeting OLFML2A (shOLFML2A) significantly inhibited cell proliferation in renal cell carcinoma (15). OLFML2A is a key promotor of gliomagenesis, and OLFML2A knockdown in glioma cells inhibited cell proliferation and promoted apoptosis through Wnt/β-catenin pathway (24). Additionally, an elevated level of OLFML2A is unfavorable for breast cancer patients according to data from The Cancer Genome Atlas (TCGA) breast cancer samples and The Human Protein Atlas (25). The overall survival rate of these breast cancer patients with different OLFML2A mRNA levels was significantly different (26). Patients with a high mRNA level of OLFML2A received a worse prognosis than those with weak and moderate mRNA levels of OLFML2A (27). Therefore, we speculate that inhibiting the expression of this gene may be beneficial to breast cancer patients. Indeed, we recently found that the inhibition of OLFML2A gene by mRNA interference markedly inhibited proliferation and migration in triple-negative breast cancer (TNBC) cells (28). However, the mechanism of inhibition of shOLFML2A in breast cancer is still not clear. Therefore, the aims of this study were to investigate the mechanism of shOLFML2A in breast cancer using a GeneChip array, RT-qPCR, and western blot analysis. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-757/rc).
Methods
Materials
An Affymetrix GeneChip Human Transcriptome Array 2.0 was obtained from Affymetrix (San Francisco, CA, USA). GeneChip hybridization, wash, and stain kits were obtained from Thermo Fisher (Waltham, MA, USA). An RNA 6000 Nano Kit was obtained from Agilent Technologies (Santa Clara, CA, USA). Dulbecco’s Modified Eagle Medium (DMEM) and fetal bovine serum (FBS) were obtained from Gibco (Grand Island, NY, USA). A mouse anti-Flag antibody (F1804) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Mouse anti-GAPDH (sc-32233), goat anti-mouse IgG (sc-2005), and anti-rabbit IgG (sc-2004) antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Rabbit anti-MDM2 (AB38618), mouse anti-SKP2 (AB183039), rabbit anti-TFDP1 (AB124678), and rabbit anti-VIMENTIN (AB92547) antibodies were purchased from Abcam (Cambridge, MA, USA). Rabbit anti-APP (#2452), rabbit anti-CCNB1 (#4138), rabbit anti-snail (#3879), and rabbit anti-slug (#9585) antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). SYBR master mix was purchased from Takara (Otsu, Shiga, Japan). TIANgel Midi Purification Kits and Endofree Maxi Plasmid Kits were obtained from Tiangen (Beijing, China). EcoRI, CutSmart and AgeI were obtained from New England Biolabs (Beverly, MA, USA). Wounding replicators were obtained from V&P Scientific (San Diego, CA, USA). dsDNA oligonucleotides, PCR primers, and a 250 bp-II DNA Ladder were purchased from Generay Biotech (Shanghai, China). Taq plus DNA polymerase was obtained from Vazyme Biotech (Nanjing, China). A Bulge-Loop miRNA qPCR primer set was obtained from RiboBio Biotech (Guangzhou, China). TRIzol, trypsin, NaCl, Tris, EDTA, and oligo dT were purchased from Sangon Biotech (Shanghai, China). DNA sequencing was performed by Majorbio Biotech (Shanghai, China).
Cells
Human breast cancer cell line MDA-MB-231 was obtained from the Shanghai Institute of Cell Biology (Shanghai, China) and cultured in DMEM supplemented with 10% FBS at 37 ℃ in a cell incubator (Thermo Fisher, MA, USA) containing 5% CO2. Green fluorescent protein (GFP)-positive cells were obtained by lentivirus particle transfection according to our previous work (28). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the First Affiliated Hospital of Zhejiang Chinese Medical University (Approval No. 2016-KL-020-02). Informed consent was obtained from all female TNBC patients.
RNA interference (RNAi) using shRNAs
To reduce OLFML2A gene expression in TNBC cells, shRNA interference was performed according to our previous work (28). Plasmid GV115 was used for lentivirus vector construction. The element sequence is hU6-MCS-CMV-EGFP. The control insert is TTCTCCGAACGTGTCACGT. The shRNA sequences are shown in Table 1. Cells were cultured in a 12-well plate at a density of 50,000 cells/well. Cells were used for subsequent experiments when more than 90% of the cells were GFP-positive.
Table 1
| Group | Target sequence | GC content |
|---|---|---|
| psc45859 | CTGGAATATACACCACTTGAA | 38.1% |
| psc45860 | GATCTATGTCACCAACTACTA | 38.1% |
| psc45861 | CTTCACCAAGAACATCATCAA | 38.1% |
RT-qPCR
Total RNA was extracted using the TRIzol reagent, and RT-qPCR was performed according to our previous work (28). Primers were obtained from GeneChem (Shanghai, China), and their sequences were as follows: 5'-TGACTTCAACAGCGACACCCA-3' (sense) and 5'-CACCCTGTTGCTGTAGCCAAA-3' (antisense) for GAPDH; and 5'-AACAGGCAGTAGAGTCAA-3' (sense), and 5'-TTACAAGATTCCTACCAACAG-3' (antisense) for OLFML2A. The relative quantitative analysis of mRNA levels was performed using the 2−ΔΔCt method (29).
Western blot analysis
Western blot analysis was performed as described in our previous work (30). The concentration of the separating gel was 10%. The amount of protein loaded was 30 μg. The blots were developed with a Pierce ECL Western Blotting Substrate Kit (Thermo Fisher, Waltham, MA, USA) and exposed on X-ray film.
Wound-healing assay
A wound-healing assay was performed according to our previous work (28). Briefly, cells were seeded into 96-well plates at a density of 50,000 cells/well in DMEM supplemented with 10% FBS. Scratches were produced with a 96-wounding replicator when cell confluence reached approximately 90%. Cells were cultured in medium containing 1% FBS, and free-floating cells were removed. Then, cells were photographed with a Celigo Imaging Cytometer (Nexcelom Bioscience, Lawrence, MA, USA). The migration rate and migration inhibition rate were calculated as described in our previous work (28).
Transwell migration assay
Transwell migration assays were performed as described previously, with slight modifications (31). MDA-MB-231 cells were seeded into 24-well transwell chambers (Corning, Corning, NY, USA) at a density of 70,000 cells/well. One hundred microliters of serum-free medium were added to the upper chamber, and 600 μL of DMEM containing 10% FBS was added to the bottom chamber. Cells were cultured for 24 h. Non-migrated cells were scraped off, and the migrated cells were stained with Giemsa and photographed with an inverted microscope.
Transwell invasion assay
Transwell invasion assays were performed as described in our previous study, with slight modifications (28). Briefly, 1×106 cells were seeded into the upper chamber of a 24-well transwell chamber separated by a polycarbonate filter coated with 50 μg/mL collagen IV and cultured with 500 μL of serum-free medium. A total of 750 μL of DMEM containing 10% FBS was supplied to the bottom chamber. After 24 h of culture, noninvasive cells were removed, and invasive cells were stained with Giemsa and photographed with an inverted microscope according to our previous work (28).
Cell proliferation analysis by the Celigo assay
Cell proliferation was analyzed using the Celigo assay as described in our previous work (28). Cells were cultured in 96-well plates at a density of 2,000 cells/well. The cells were photographed with Celigo, and a cell growth curve was drawn to reflect cell proliferation following 5 days of consecutive recording.
Cell activity analysis by the MTT assay
Cell activity was assessed using the MTT method according to our previous work (32). Briefly, cells were cultured in a 96-well plate at a density of 2,000 cells/well. Cell activity was determined with a microplate reader on 5 consecutive days using MTT methods.
Immunohistochemical (IHC) analysis
IHC analysis was carried out according to previous work (33). The dilution ratio of the antibody was in accordance with the instructions provided by the antibody company.
GeneChip microarray
Total RNA was extracted using the TRIzol method and examined on a NanoDrop 2000 Spectrophotometer (Thermo Scientific, MA, USA) and an Agilent Bioanalyzer 2100 (Palo Alto, CA, USA) as described in our previous work (28). Only qualified samples were used in subsequent microarray experiments. The GeneChip was processed as described in our previous work (28). Background noise was filtered from the two groups within the lowest 20% range of the sequence of signal strength of all probe groups. The probe with a coefficient of variation >20% in the same group was also removed (34). The number of initial probes was 49,395. Finally, there were 39,260 probes after filtering. Genes that were up- or downregulated with the |fold changes| >2.0.
Ingenuity pathway analysis (IPA)
The IPA of differentially expressed genes (DEGs) was using Qiagen’s Ingenuity Pathway Analysis algorithm (www.qiagen.com/ingenuity, Qiagen, Redwood City, CA, USA). The activation z-score and P value were calculated according to previous work (35).
Statistical analysis
All results are presented as the mean ± standard deviation. Two-tailed analysis of variance (ANOVA) followed by Dunnett’s post-hoc test was performed to determine statistical significance. For the microarray, a linear model based on the empirical Bayesian distribution was used to calculate the P value of the significant difference level, and the Benjamini-Hochberg method was used to control the false discovery rate (FDR). The screening criteria for significant differentially expressed genes were |fold change| >2 and FDR <0.05.
Results
shOLFML2A reduces breast cancer cell malignancy
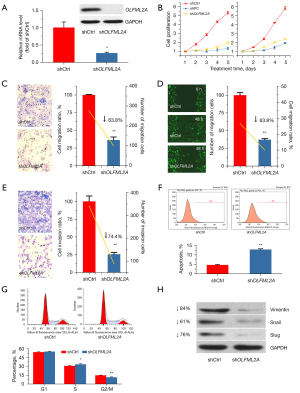
To increase the efficiency of RNAi, three shRNAs were designed for shOLFML2A, and their sequences are shown in Table 1. Cell proliferation and migration assays were performed to compare the effectiveness of these shRNAs after the breast cancer cell line MBA-MD-231 was transfected with these target plasmids. All three shOLFML2As significantly inhibited cell proliferation (Figure S1A-S1C). In addition, shOLFML2A-psc45860 and shOLFML2A-psc45861 also decreased cell migration (Figure S1D-S1F). Therefore, shOLFML2A-psc45860 (shOLFML2A) was selected for subsequent experiments. Then, we verified the OLFML2A gene expression in the gene-silenced cell through RT-qPCR and western blot analysis. Compared to its wild-type control, shOLFML2A significantly decreased OLFML2A mRNA levels and inhibited OLFML2A protein expression (Figure 1A). Celigo and MTT assays revealed that shOLFML2A significantly inhibited tumor cell proliferation and viability (Figure 1B). The transwell migration assay and wound-healing analysis revealed that shOLFML2A decreased cancer cell migration (Figure 1C,1D). shOLFML2A also reduced cell invasion (Figure 1E). We subsequently evaluated the changes in the cell cycle and apoptosis between OLFML2A gene-silenced cells and wild-type control cells. shOLFML2A induced cell apoptosis by promoting S phase arrest in breast cancer cells (Figure 1F,1G). In addition, shOLFML2A decreased the protein levels of typical epithelial-mesenchymal transition (EMT) markers, including vimentin, snail, and slug (Figure 1H).
Gene alterations by shOLFML2A in breast cancer cells
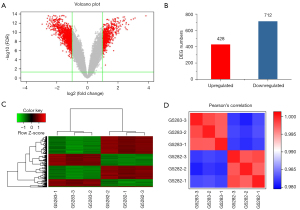
We then determined the genome-wide expression changes between OLFML2A gene-silenced cells and wild-type control cells in MBA-MD-231 breast cancer cells using microarrays. A large number of genes were significantly altered by shOLFML2A in breast cancer cells according to the volcano plots (Figure 2A) and scatter plots of the microarrays (Figure S2). Among the DEGs (|fold change| >2 and FDR <0.05), 428 were upregulated and 712 were downregulated (Figure 2B). A heatmap of the microarray created upon comparison of the DEGs of all samples showed gene clustering (Figure 2C). The relationships of these DEGs are shown in the website: https://cdn.amegroups.cn/static/public/atm-22-757-01.pdf. Many of these genes play key roles in cancer, cell proliferation, the cell cycle, and cellular movement, and details can be found in the website: https://cdn.amegroups.cn/static/public/atm-22-757-02.xlsx. Pearson’s correlation coefficient of the RNA samples among the gene-silenced cells or the wild-type control cells was >0.95, suggesting that the microarray results were reliable (Figure 2D). Principal component analysis (PCA) also indicated that the samples in the same group were similar and that the difference between groups was different (Figure S2A). The relative signal box plot array indicated that the microarray data were reproducible (Figure S2C).
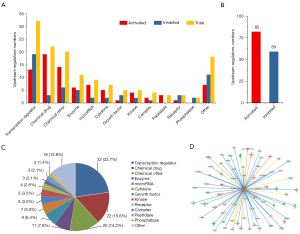
Changes in canonical pathways by shOLFML2A
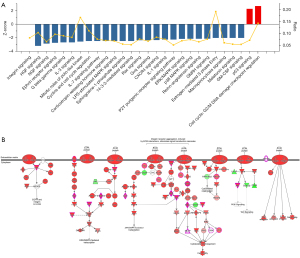
Based on the DEGs, an IPA was performed to identify the canonical pathways of the DEGs. There were 127 pathways changed by shOLFML2A in breast cancer cells (available online: https://cdn.amegroups.cn/static/public/atm-22-757-03.xlsx). Among them, 27 tumor-associated signaling pathways changed significantly (Figure 3A). The ratio of the DEGs to the total genes in the canonical pathways varied from 5.2% to 19.2% (Figure 3A). shOLFML2A activated the cell cycle: G2/M DNA damage checkpoint regulation and p53 signaling pathways. The other 25 pathways, including integrin signaling, HGF signaling, and NGF signaling, were significantly inhibited by shOLFML2A. Integrin signaling, which induces ECM interactions and stimulates signal transduction cascades, was found to be the top pathway (Figure 3B). The integrin signaling pathway regulates cell motility, cell adhesion, cell activation, and tumor invasion through crosstalk with the EGFR, PI3K/AKT, ERK/MAPK, JNK/SAPK, Wnt and cytoskeletal organization/rearrangement signaling pathways (Figure 3B). AKT, CAV1, GSN, MLCP, PAK, PINCH1, RAP, Ras, RHO, TSPAN, VCL, and WASP are involved in the suppression of integrin signaling (Figure 3B). AKT3, CAV1, ELK1, PAK1, PTK2, RAC1, RAP1A/B, and WASL participates in the regulation of the top 6 signaling pathways, namely, integrin signaling, HGF signaling, NGF signaling, ephrin receptor signaling, G beta gamma signaling, and the IL-3 signaling pathway (Figure S3A). The other signaling pathways and their molecules can be found in the website: https://cdn.amegroups.cn/static/public/atm-22-757-03.xlsx. Among them, Prkce, Prkch, Akt3, Rac1, Elk1, Ptk2, Pak1, Rhoa, and Prkacb were the principal molecules involved in these significantly changed pathways (Figure S3B).
Functions of the DEGs by shOLFML2A
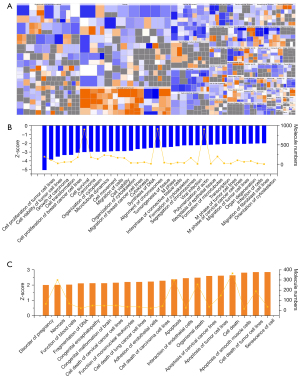
We then further analyzed the functions of these differential canonical pathways regulated by shOLFML2A in breast cancer cells by IPA. More than 500 functions were changed by shOLFML2A, 54 of which were altered significantly (Figure 4 and available online: https://cdn.amegroups.cn/static/public/atm-22-757-04.xlsx). Among the significantly changed functions, 34 were significantly inhibited, and 20 were activated (Figure 4). Among them, the synthesis of DNA, the alignment of chromosomes, the formation of microtubules and the cytoskeleton, cell viability, cell proliferation, cell movement and migration, etc. were inhibited (Figure 4B). However, necrosis, cell death, apoptosis, senescence, cell adhesion, and the interaction of endothelial cells were activated (Figure 4C). The numbers of molecules engaged in these functions ranged from 4 to 911 (Figure 4B,4C). The top function was the cell proliferation of tumor cell lines (Figure S4A). There were 123 inhibited molecules and 74 activated molecules in this function (Figure S4B). The other functions and their molecules can be found in the available online: https://cdn.amegroups.cn/static/public/atm-22-757-04.xlsx. Among them, Rac1, Rhoa, App, Bcl2, Bcl2l1, Birc5, Il1b, Vegfa, Pak1 and Cav1 were the top molecules involved in these significantly changed functions (Figure S4C).
Upstream regulators of the DEGs by shOLFML2A
The upstream regulators of the DEGs by shOLFML2A were analyzed by IPA. There were 2,260 regulators that were significantly changed by shOLFML2A in breast cancer cells (available online: https://cdn.amegroups.cn/static/public/atm-22-757-05.xlsx). Among them, there were 141 with an |activation z-score| >2 (Figure 5A,5B). Transcription regulators were the top regulators according to the classification analysis (Figure 5C). The top regulator was the transcription regulator NUPR1, as shown in Figure 5D. Additionally, NUPR1 could activate 14 target molecules and inhibit 56 molecules in the dataset (Figure S5A). The top 10 inhibited upstream regulators and their representative targets, such as ESR1, RABL6, and FoXM1, are shown in Figure S5B. The top 10 activated regulators (e.g., NUPR1, tretinoin, and let-7) are shown in Figure S5C. For more detailed information, please refer to the available online: https://cdn.amegroups.cn/static/public/atm-22-757-05.xlsx.
Regulator effects and interaction networks of the DEGs by shOLFML2A
IPA was further performed to explore the relationships between the upstream regulators and the downstream functions of shOLFML2A in breast cancer cells. A higher consistency score indicates a more accurate prediction of these relationships. The top relationship was the adhesion of endothelial cells and increased levels of albumin (Figure 6A). Among them, ADM, Ap1, ATF6, CYP2J2, HMOX1, IL1RN, NOX4, TGM2, and VCAN regulate APP, FN1, ICAM1, IL1B, PPARG, PROCR, RCAN1, and VEGFA expression to regulate cell adhesion and increase albumin. Details can be found in Figure S6A and the available online: https://cdn.amegroups.cn/static/public/atm-22-757-06.xlsx. The top 5 regulators play significant roles in cell apoptosis, cell death, cell survival, DNA fragmentation, and microtubule dynamics (Figure S6A).
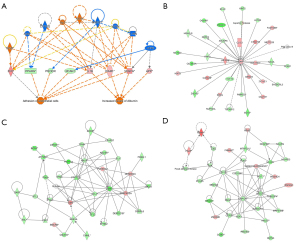
We then constructed the interaction network among genes in the dataset, upstream regulators, and cell functions using IPA. The overall network of molecules was divided into multiple networks, and then each network was scored. A higher score indicates a higher probability prediction of these interactions. Three networks topped the rankings jointly with a score of 45 (Figure 6B-6D). APP, PI3K, PTK, etc. participate in the regulation of DNA replication, recombination, and repair, the cell cycle, cellular assembly and organization, cellular compromise, cellular assembly and organization, and cellular development (Figure S6B). For more information, please refer to the available online: https://cdn.amegroups.cn/static/public/atm-22-757-07.xlsx.
Confirmation of the microarray by RT-qPCR, western blot and IHC analyses
To verify the reliability of the microarray, RT-qPCR and western blot analysis were employed. All the candidate genes that encoded multiple transmembrane proteins and those that were not well annotated in the National Center for Biotechnology Information (NCBI) database were not included in this study. Finally, 30 genes were chosen for subsequent confirmatory experiments. APP, EGR1, GDG15, HDAC6, MMP1, and TNFSF10 were upregulated by shOLFML2A in TNBC cells; however, CD/CDCs, RAC1, RHOA, etc. were downregulated compared to their parental cells (Figure 7A). The RT-qPCR results were also similar to those of the microarray (Figure 7B). Correlation analysis showed that the microarray and RT-qPCR results were highly correlated, with a correlation coefficient R2>0.9 (Figure 7C). Additionally, the protein expression of some genes was analyzed. shOLFML2A significantly decreased the protein expression of CCNB1, MDM2, OLFML2A, SKP2, and TFDP1 and increased the protein expression of APP (Figure 7D). The IHC results indicated that expression of OLFML2A was increased in TNBC cells compared to the paracancerous tissue (Figure 7E).
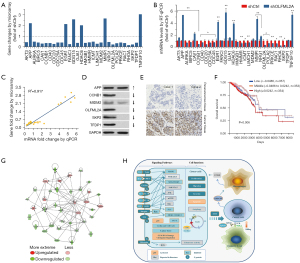
Gene relation network by shOLFML2A in TNBC cells
In humans, the OLFML2A gene is expressed in the breast, cerebral cortex, colon, eye, hippocampus, kidney, etc. under physiological conditions according to data from The Human Protein Atlas (Figure S7A,S7B) (4). Its protein can be detected in several human neoplastic tissues, such as breast, cervical, colorectal, endometrial, liver, lung, ovarian, pancreatic, and stomach cancers (Figure S7C) (14). According to data from the TCGA breast cancer samples and The Human Protein Atlas, a high level of OLFML2A is unfavorable (Figure S7D,S7E) (24). The overall survival rate of female breast cancer patients with different levels of OLFML2A gene expression was significantly different (Figure 7F). Patients with a high level of OLFML2A gene expression received a worse prognosis than those with weak and moderate levels of OLFML2A gene expression (Figure 7F). A gene relation network was eventually obtained by the intersection analysis of the DEGs involved in DNA replication, recombination and repair, the cell cycle, cellular assembly and organization, cell proliferation, cell death and apoptosis, cell movement and migration, cell cycle regulation, integrin signaling, HGF signaling, and NGF signaling (Figure 7G). In breast cancer MBA-MD-231 cells, shOLFML2A downregulated AKT3, AURKA, BIRC5, CAV1, CCNB1, CD40, CDC20, CDC25A, CDC45, DEF6, FANCI, GJA1, HMGB1, HMMR, HSPA8, ICMT, LIMCH1, LPAR1, MDM2, NRG1, OLFML2A, POLA2, PRKCE, RAC1, RHOA, SKP2, and TFDP1 and upregulated APP, EGR1, GDF15, GRB10, HDAC6, MMP1, MMP3, SQSTM1, TNFSF10, and TSC22D1. For details, please refer to the available online: https://cdn.amegroups.cn/static/public/atm-22-757-08.xlsx. Lastly, we drew a schematic diagram of the mechanism of the inhibitory effects of shOLFML2A in breast cancer cells (Figure 7H). In human breast cancer MBA-MD-231 cells, gene silencing of OLFML2A inhibited cell proliferation, migration, and invasion and promoted cell apoptosis potentially through signaling crosstalk with the p53, integrin, HGF, NGF, ephrin receptor, Gβγ, IL-3, mitotic roles of polo-like kinase, cell cycle regulation, IL-7, corticotropin releasing hormone, sphingosine-1-phosphate, 14-3-3-mediated, Rac, Gαq, CXCR4, IL-1, P2Y purigenic receptor, ERK/MAPK, p38 MAPK, renin-angiotensin, gonadotropin-releasing hormone (GNRH), melatonin, and granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling pathways.
Discussion
In this study, we investigated the mechanisms of the inhibitory action of OLFML2A silencing using RNAi on human breast cancer cells. High OLFML2A expression is unfavorable in breast cancer and is a potential prognostic factor. In breast cancer MBA-MD-231 cells, shOLFML2A inhibited cell proliferation and viability, cell migration and invasion, and the EMT progress because it induced cell apoptosis by promoting S phase arrest. It also inhibited cell functions, such as the synthesis of DNA, the alignment of chromosomes, and the formation of microtubules, and the cytoskeleton, while cell necrosis, cell death, apoptosis, senescence, cell adhesion, and the interaction of endothelial cells were activated. In the cell cycle, G2/M DNA damage checkpoint regulation and p53 signaling were activated by shOLFML2A. However, shOLFML2A significantly inhibited tumor-promoting signaling pathways, such as integrin, HGF, NGF, ephrin receptor, Gβγ, IL-1/3/7, the mitotic roles of polo-like kinase, cyclins and cell cycle regulation, sphingosine-1-phosphate, CXCR4, ERK/MAPK, p38 MAPK, melatonin, GM-CSF, 14-3-3, corticotropin-releasing hormone, Rac, P2Y purigenic receptor, renin-angiotensin, GNRH, estrogen-mediated S phase entry, and macropinocytosis signaling. Therefore, OLFML2A was revealed as a potential therapeutic target in TNBC cells.
OLFML2A protein is an abnormal protein that is widely distributed in human neoplastic tissues, such as breast, cervical, colorectal, endometrial, liver, lung, ovarian, pancreatic, and stomach cancers (4). We also found elevated OLFML2A expression in some breast cancer patients. Additionally, a high level of OLFML2A is unfavorable for female breast cancer patients according to data from TCGA breast cancer samples and The Human Protein Atlas (25). The overall survival rate of these patients with different OLFML2A mRNA levels was significantly different, with a P value of 0.006 (25). Patients with a high mRNA level of OLFML2A received a worse prognosis than those with weak and moderate mRNA levels of OLFML2A. Therefore, we speculate that inhibiting the expression of this gene may be beneficial to breast cancer patients. Indeed, we recently found that silencing of the OLFML2A gene by mRNA interference significantly inhibited proliferation and migration in TNBC cells (28). Therefore, we investigated the mechanism of the inhibition of shOLFML2A by using a GeneChip array, RT-qPCR, m and western blot analysis. In the present investigation, we further found that shOLFML2A induced cell apoptosis by promoting S phase arrest in breast cancer cells. shOLFML2A also decreased the protein expression of several typical EMT markers, such as vimentin, snail, and slug. Cell cycle regulation and the apoptosis-promoting effect of shOLFML2A may decrease cell proliferation, migration, and invasion.
To explore the genes that were significantly changed by shOLFML2A, we performed a microarray and found that 428 upregulated and 712 downregulated genes were changed by shOLFML2A in breast cancer cells; thirty were tested by RT-qPCR. The microarray and RT-qPCR results were highly correlated. All these significantly changed genes were further analyzed using IPA. Interestingly, shOLFML2A activated G2/M DNA damage checkpoint regulation and the p53 signaling pathways. The activation of these pathways is beneficial to the prevention and treatment of breast cancer (36,37). shOLFML2A also inhibited tumor-promoting signaling pathways, such as integrin, HGF, NGF, ephrin receptor, Gβγ, IL-1/3/7, the mitotic roles of polo-like kinase, cyclins and cell cycle regulation, sphingosine-1-phosphate, CXCR4, ERK/MAPK, p38 MAPK, melatonin, GM-CSF, 14-3-3-mediated signaling, corticotropin-releasing hormone, Rac, P2Y purigenic receptor, renin-angiotensin, GNRH, estrogen-mediated S phase entry, and macropinocytosis signaling. In particular, integrin signaling was the top pathway, and it induced ECM interactions. The integrin signaling pathway regulates cell motility, cell adhesion, cell activation, and tumor invasion through signaling crosstalk with the EGFR, PI3K/AKT, ERK/MAPK, JNK/STPK, Wnt/β-catenin, and cytoskeletal organization/rearrangement signaling pathways (38,39). Breast cancer cells exhibit a metastatic phenotype that is controlled by the activation of integrin signaling, andinhibition of the integrin pathway decreases tumor metastasis(40). In addition, the high-frequency molecules Prkce, Prkch, Akt3, Rac1, Elk1, Ptk2, Rhoa, Pak1/6, and Prkacb were observed in the signaling pathways that were significantly changed by shOLFML2A. These genes play key roles in breast cancer tumorigenesis (41-43).
We then further analyzed the functional changes induced by shOLFML2A using IPA in breast cancer cells. Among the significantly changed functions, 34 were significantly inhibited and 20 were activated by shOLFML2A. Surprisingly, shOLFML2A inhibited the synthesis of DNA, the alignment of chromosomes, the formation of microtubules and the cytoskeleton, cell viability, cell proliferation, and cell movement and migration but promoted necrosis, cell death, apoptosis, senescence, cell adhesion, and the interaction of endothelial cells. This evidence indicates that silencing of the OLFML2A gene decreases tumorigenesis and tumor progression in breast cancer. Additionally, the high-frequency molecules Rac1, Rhoa, Bcl2, and APP are involved in these functions. We further explored the relationships between the upstream regulators and the downstream functions changed by shOLFML2A. The adhesion of endothelial cells and increased levels of albumin were the top relationship. ADM, Ap1, ATF6, CYP2J2, HMOX1, IL1RN, NOX4, TGM2, and VCAN regulated APP, FN1, ICAM1, IL1B, PPARG, PROCR, RCAN1, and VEGFA expression to regulate cell adhesion and increase albumin. The regulation of cell motility and cell adhesion, which is controlled by microtubules and the cytoskeleton, is critical to cancer cell metastases (44). Additionally, attenuation of the adhesion of tumor cells to endothelial cells is beneficial for breast cancer cell metastasis (45). The top 5 regulator effects play significant roles in cell apoptosis, cell death, cell survival, DNA fragmentation, and microtubule dynamics. These data indicate that the regulation of DNA synthesis and microtubule dynamics by shOLFML2A reduces proliferation and adhesion in breast cancer cells.
Conclusions
Based on the GeneChip and IPA results, we further analyzed the core interaction of the genes involved in the functions of cancer, cell proliferation, the cell cycle, apoptosis, and cellular movement. APP, RAC1, CAV1, HSPA8, AURKA, CDC20, HDAC6, CCNB1, GJA1, and SQSTM1 constituted the core of this network. Taken together, these results suggest that gene silencing by shOLFML2A changes the expression of APP, RAC1, CAV1, and other genes. These changed genes then regulate DNA synthesis, chromosome alignment, microtubules and the cytoskeleton, cell movement, the cell cycle, cell necrosis, and apoptosis by promoting regulation of the G2/M DNA damage checkpoint and p53 signaling and by inhibiting integrin, HGF, NGF, and other tumor-promoting signaling pathways. Finally, shOLFML2A reduced cell proliferation, migration, and invasion and promoted cell apoptosis. Therefore, the results of the present investigation suggest that OLFML2A is a potential therapeutic target for TNBC cells.
Acknowledgments
Funding: This research was supported by The National Natural Science Foundation of China (No. 82074438), the Natural Science Foundation of Zhejiang Province (No. LY18H270006), and the Zhejiang Province Traditional Chinese Medicine Scientific Research Foundation (No. 2016ZQ015).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-757/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-757/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-757/coif). All authors report that this work was supported by The National Natural Science Foundation of China (No. 82074438), the Natural Science Foundation of Zhejiang Province (No. LY18H270006), and the Zhejiang Province Traditional Chinese Medicine Scientific Research Foundation (No. 2016ZQ015). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the First Affiliated Hospital of Zhejiang Chinese Medical University (Approval No. 2016-KL-020-02). Informed consent was obtained from all female TNBC patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mukhopadhyay A, Talukdar S, Bhattacharjee A, et al. Bioinformatic approaches for identification and characterization of olfactomedin related genes with a potential role in pathogenesis of ocular disorders. Mol Vis 2004;10:304-14. [PubMed]
- Tomarev SI, Nakaya N. Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Mol Neurobiol 2009;40:122-38. [Crossref] [PubMed]
- Furutani Y, Manabe R, Tsutsui K, et al. Identification and characterization of photomedins: novel olfactomedin-domain-containing proteins with chondroitin sulphate-E-binding activity. Biochem J 2005;389:675-84. [Crossref] [PubMed]
- Available online: https://www.proteinatlas.org/ENSG00000185585-OLFML2A/tissue.
- Pérez-Ibave DC, González-Alvarez R, de La Luz Martinez-Fierro M, et al. Olfactomedin-like 2 A and B (OLFML2A and OLFML2B) expression profile in primates (human and baboon). Biol Res 2016;49:44. [Crossref] [PubMed]
- Sistani L, Rodriguez PQ, Hultenby K, et al. Neuronal proteins are novel components of podocyte major processes and their expression in glomerular crescents supports their role in crescent formation. Kidney Int 2013;83:63-71. [Crossref] [PubMed]
- Yan W, Zhao Z, Zhang L, et al. Identification of estrogen-associated intrinsic aging genes in Chinese Han female skin by cDNA microarray technology. Biomed Environ Sci 2011;24:364-73. [PubMed]
- Schubert AK, Smink JJ, Arp M, et al. Quality assessment of surgical disc samples discriminates human annulus fibrosus and nucleus pulposus on tissue and molecular level. Int J Mol Sci 2018;19:1761. [Crossref] [PubMed]
- Zhao X, Liang M, Li X, et al. Identification of key genes and pathways associated with osteogenic differentiation of adipose stem cells. J Cell Physiol 2018;233:9777-85. [Crossref] [PubMed]
- Kim JM, Yu JM, Bae YC, et al. Analysis of global gene expression profile of human adipose tissue derived mesenchymal stem cell cultured with cancer cells. J Life Sci 2011;21:631-46. [Crossref]
- Jiang XY, Ning QL. Expression profiling of long noncoding RNAs and the dynamic changes of lncRNA-NR024118 and Cdkn1c in angiotensin II-treated cardiac fibroblasts. Int J Clin Exp Pathol 2014;7:1325-36. [PubMed]
- McGaugh SE, Gross JB, Aken B, et al. The cavefish genome reveals candidate genes for eye loss. Nat Commun 2014;5:5307. [Crossref] [PubMed]
- Lee JA, Anholt RR, Cole GJ. Olfactomedin-2 mediates development of the anterior central nervous system and head structures in zebrafish. Mech Dev 2008;125:167-81. [Crossref] [PubMed]
- Available online: https://www.proteinatlas.org/ENSG00000185585-OLFML2A/pathology.
- Von Roemeling CA, Marlow LA, Radisky DC, et al. Functional genomics identifies novel genes essential for clear cell renal cell carcinoma tumor cell proliferation and migration. Oncotarget 2014;5:5320-34. [Crossref] [PubMed]
- Ramakrishnan S, Huss W, Foster B, et al. Transcriptional changes associated with in vivo growth of muscle-invasive bladder cancer cell lines in nude mice. Am J Clin Exp Urol 2018;6:138-48. [PubMed]
- Kobos R, Nagai M, Tsuda M, et al. Combining integrated genomics and functional genomics to dissect the biology of a cancer-associated, aberrant transcription factor, the ASPSCR1-TFE3 fusion oncoprotein. J Pathol 2013;229:743-54. [Crossref] [PubMed]
- Hayase S, Sasaki Y, Matsubara T, et al. Expression of stanniocalcin 1 in thyroid side population cells and thyroid cancer cells. Thyroid 2015;25:425-36. [Crossref] [PubMed]
- Wang S, Zhang L, Shi P, et al. Genome-wide profiles of metastasis-associated mRNAs and microRNAs in salivary adenoid cystic carcinoma. Biochem Biophys Res Commun 2018;500:632-8. [Crossref] [PubMed]
- López J, Poitevin A, Mendoza-Martínez V, et al. Cancer-initiating cells derived from established cervical cell lines exhibit stem-cell markers and increased radioresistance. BMC Cancer 2012;12:48. [Crossref] [PubMed]
- Boren T, Xiong Y, Hakam A, et al. MicroRNAs and their target messenger RNAs associated with endometrial carcinogenesis. Gynecol Oncol 2008;110:206-15. [Crossref] [PubMed]
- Wu X. Up-regulation of YPEL1 and YPEL5 and down-regulation of ITGA2 in erlotinib-treated EGFR-mutant non-small cell lung cancer: A bioinformatic analysis. Gene 2018;643:74-82. [Crossref] [PubMed]
- Peng D. The effect of knockdown OLFML2A gene on the biological behavior of hepatocellular carcinoma cells. Ph.D Thesis. Nanchang: Nanchang University. 2018.
- Ma S, Duan L, Dong H, et al. OLFML2A downregulation inhibits glioma proliferation through suppression of Wnt/β-catenin signaling. Front Oncol 2021;11:717917. [Crossref] [PubMed]
- Available online: https://www.proteinatlas.org/ENSG00000185585-OLFML2A/tissue#gene_information.
- Available online: https://xenabrowser.net/heatmap/.
- Zhao Q, Zhang K, Li Y, et al. OLFML2A is necessary for anti-triple negative breast cancer effect of selective activator protein-1 inhibitor T-5224. Transl Oncol 2021;14:101100. [Crossref] [PubMed]
- Wu C, Chen M, Zhang Q, et al. Genomic and GeneChip expression profiling reveals the inhibitory effects of Amorphophalli Rhizoma in TNBC cells. J Ethnopharmacol 2019;235:206-18. [Crossref] [PubMed]
- Kubista M, Andrade JM, Bengtsson M, et al. The real-time polymerase chain reaction. Mol Aspects Med 2006;27:95-125. [Crossref] [PubMed]
- Wu C, Sun Z, Guo B, et al. Osthole inhibits bone metastasis of breast cancer. Oncotarget 2017;8:58480-93. [Crossref] [PubMed]
- Wu C, Qiu S, Liu P, et al. Rhizoma Amorphophalli inhibits TNBC cell proliferation, migration, invasion and metastasis through the PI3K/Akt/mTOR pathway. J Ethnopharmacol 2018;211:89-100. [Crossref] [PubMed]
- Chen MC, Ye YY, Ji G, et al. Hesperidin upregulates heme oxygenase-1 to attenuate hydrogen peroxide-induced cell damage in hepatic L02 cells. J Agric Food Chem 2010;58:3330-5. [Crossref] [PubMed]
- Shi SR, Liu C, Young L, et al. Development of an optimal antigen retrieval protocol for immunohistochemistry of retinoblastoma protein (pRB) in formalin fixed, paraffin sections based on comparison of different methods. Biotech Histochem 2007;82:301-9. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Krämer A, Green J, Pollard J Jr, et al. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30:523-30. [Crossref] [PubMed]
- Cuddihy AR, O'Connell MJ. Cell-cycle responses to DNA damage in G2. Int Rev Cytol 2003;222:99-140. [Crossref] [PubMed]
- Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A 2005;102:13550-5. [Crossref] [PubMed]
- Levental KR, Yu H, Kass L, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 2009;139:891-906. [Crossref] [PubMed]
- Chan SH, Huang WC, Chang JW, et al. MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene 2014;33:4496-507. [Crossref] [PubMed]
- Felding-Habermann B, O'Toole TE, Smith JW, et al. Integrin activation controls metastasis in human breast cancer. Proc Natl Acad Sci U S A 2001;98:1853-8. [Crossref] [PubMed]
- Dann SG, Golas J, Miranda M, et al. p120 catenin is a key effector of a Ras-PKCɛ oncogenic signaling axis. Oncogene 2014;33:1385-94. [Crossref] [PubMed]
- Mosquera JM, Varma S, Pauli C, et al. MAGI3-AKT3 fusion in breast cancer amended. Nature 2015;520:E11-2. [Crossref] [PubMed]
- Zhang Z, Yang M, Chen R, et al. IBP regulates epithelial-to-mesenchymal transition and the motility of breast cancer cells via Rac1, RhoA and Cdc42 signaling pathways. Oncogene 2014;33:3374-82. [Crossref] [PubMed]
- Hill A, McFarlane S, Mulligan K, et al. Cortactin underpins CD44-promoted invasion and adhesion of breast cancer cells to bone marrow endothelial cells. Oncogene 2006;25:6079-91. [Crossref] [PubMed]
- Bischofs E, Lubs D, Fritzsche F, et al. In vitro blockade of adhesion of breast cancer cells to endothelial cells using anti-inflammatory drugs. Anticancer Res 2012;32:767-71. [PubMed]


