
Acute intermittent porphyria: prevalence of pathogenic HMBS variants in China, and epidemiological survey in Hebei Province, China
Introduction
Porphyrias are a group of metabolic diseases resulting from a specific abnormality in one of the 8 enzymes in the heme biosynthetic pathway (1). Acute intermittent porphyria (AIP; Online Mendelian Inheritance in Man [OMIM]#176000), the most common and severe form of acute hepatic porphyria, is an autosomal dominant genetic disorder stemming from a variant in the gene encoding hydroxymethylbilane synthase (HMBS; EC 2.5.1.61), the third enzyme in the heme biosynthesis pathway (2,3). The pathogenesis of AIP is not completely clear. Partial defects of HMBS enzyme result in the accumulation of δ-aminolevulinic acid (ALA) and porphobilinogen (PBG), which are most likely responsible for neurotoxicity, and consequently triggers off acute episodes including abdominal pain, hypertension, neuropsychiatric symptoms, severe hyponatremia, and even life-threatening attacks (4,5). Alternately, AIP is characterized by extremely low clinical penetrance in the general population, at only approximately 1% (6,7). Acute attacks can be caused by directly or indirectly inducing δ-aminolevulinic acid synthase (ALAS; EC 2.3.1.37)1(ALAS1), the rate-limiting enzyme of heme biosynthetic pathway in liver (8). Alcohol, smoking, drugs, fasting/hunger, and changes in women’s sex hormone levels are the common precipitants (1,9). Differences have been found in the serum metabolic profiles between normal individuals and patients carrying the HMBS variant (10). All individuals with pathogenic HMBS gene variants are at risk of developing acute attacks, and chronic complications such as hepatocellular carcinoma, hypertension, and chronic renal failure can occur in both symptomatic AIP and latent patients with pathogenic HMBS variants (8,11). Thus, understanding the prevalence of pathogenic HMBS variants in the general population is of great significance for early detection of potential patients and improvement of clinical prognosis. The sliding prevalence of symptomatic AIP in some countries, such as Finland, has indicated the importance of improved management and educational strategies (12).
To date, although some reports have studied the variants of pathogenic HMBS in symptomatic AIP patients in China, both the prevalence of pathogenic HMBS variants in the general Chinese population and epidemiological characteristics of AIP patients in China are still lacking.
So far, the data of prevalence of pathogenic HMBS variants have mostly come from studies in European and American populations. While, these data may not be applicable to Chinese due to ethnic differences. Here, we identified pathogenic/likely pathogenic (P/LP) HMBS variants in the China Metabolic Analysis Project database (ChinaMAP; www.mBiobank.com) according to the American College of Medical Genetics and Genomics (ACMG) guidelines to calculate the prevalence of pathogenic HMBS variants in the Chinese population. Then, to confirm the genetic heterogeneity of AIP, we predicted the prevalence in several other ethnic groups on the basis of the Genome Aggregation Database (GnomAD) Genome V3.0 (https://gnomad.broadinstitute.org/blog/2019-10-gnomad-v3-0/) database to analyze the distribution patterns of pathogenic HMBS variants among various racial populations.
The incidence of symptomatic AIP and other epidemiological characteristics have been studied in Europe (13,14), which are probably not transferrable to the Chinese population due to the obvious distinctions in ethnic characteristics. To preliminarily reveal the epidemiological characteristics of AIP in the Chinese population, we investigated the annual incidence of AIP during a 10-year period from January 2011 to December 2020 in 32 comprehensive Grade III A tertiary hospitals in Hebei province and analyzed the patients’ gender, age, and consulting departments. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1600/rc).
Methods
Database variants collection
The nomenclature of the HMBS variants was in accordance with the standards of the Human Genome Variation Society (HGVS; http://www.HGVS.org/varnomen). DNA and protein sequence numbering was completed according to the reference sequence: RefSeq NM_000190.4 and NP_000181.2, respectively.
There are magnificent databases of human genomics and bioinformatics, but none contains enough information about Chinese people before ChinaMAP database came along. ChinaMAP is based on cohort studies across diverse regions and ethnic groups with metabolic phenotypic data in China, which contains deep whole genome sequencing (WGS) data of 10,588 Chinese individuals (21,176 alleles) (15). Here, the ChinaMAP database was adopted as a surrogate for the general Chinese population (CHI). In addition, the GnomAD Genome V3.0 database, based on WGS results of approximately 143,356 alleles, was selected as the source of gene variation data for other racial general populations, including East Asian (EAS), Ashkenazi Jewish (ASJ), Mixed American (AMR), African/African American (AFR), Amish (AMI), Finnish (FIN), Non-Finnish European (NFE), South Asian (SAS), and others (OTH). The gnomAD database is the largest annotated population frequency database, with data from various disease research projects and sequencing projects in large populations.
Interpretation of variants based on the ACMG guidelines
The ACMG convened a workgroup in 2013 comprising representatives from the ACMG, the Association for Molecular Pathology (AMP), and the College of American Pathologists to revise the standards and guidelines for the interpretation of sequence variants. In accordance with the ACMG guidelines and recommendations, variants were classified as pathogenic (P), likely pathogenic (LP), benign (B), likely benign (LB), and variant of uncertain significance (VUS). The HMBS variants detected from the above databases were interpreted strictly according to the ACMG guidelines.
Variants in the exonic and splicing regions with an allele frequency of ≤0.05 in the following databases were screened out: 1000 Genomes Project, Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org/), Exome Variant Server (EVS; http://evs.gs.washington.edu/EVS), and Genome Aggregation Database (gnomAD, https://gnomad.broadinstitute.org/blog/2019-10-gnomad-v3-0/).
During this interpreting process, multiple in silico tools were selected to help predict the pathogenicity of variants. These algorithms, including SIFT, Polyphen2_HDIV, Polyphen2_HVAR, LRT, MutationTaster, MutationAssessor, FATHMM, PROVEAN, M-CAP, and CADD, were employed to predict whether a missense change was damaging to the resultant protein function or structure, and dbscSNV_ADA, dbscSNV_RF, SpliceAI, MaxEntScan, and MMSplice, which could estimate whether it exerted effects on splicing. Subsequently, rare exome variant ensemble learner (REVEL) and s-PP3 were estimated. The REVEL is a score that combines the results of multiple software predictions including SIFT and Polyphen, and PP3 evidence of the ACMG guidelines can be utilized when REVEL score is >0.75. The S-PP3 is a synthesized score of 5 splicing prediction software (dbscSNV_ADA, dbscSNV_RF, MMSplice, MaxEnt, SpliceAI), with 1 point for each piece of software that is deemed to affect the splicing.
Predicting prevalence of pathogenic HMBS variants in Chinese and other racial groups
Variants interpreted as P/LP by the ACMG guidelines were screened out to predict the prevalence of pathogenic HMBS variants. The distribution characteristics of P/LP HMBS variants and variants of unknown significance tending to pathogenic (VUS-P) (REVEL >0.7, or nonsynonymous variants that were predicted to be pathogenic by at least 3 software, or were recorded as pathogenic in the database, there was no benign evidence. Splicing variants that were considered to affect splicing by at least 2 software) among diverse racial groups were analyzed subsequently.
Epidemiological investigation of AIP in Hebei, China
Due to it being extremely rare, few physicians are able to diagnose AIP in outpatient clinics. Most patients with AIP have received no effective treatment or remission before diagnosis and need to be hospitalized. Furthermore, primary medical institutions have limited ability to diagnose AIP. Therefore, we thought it reasonable to select the cases hospitalized in comprehensive grade III A tertiary hospitals to approximately represent the number of AIP patients in a certain province. Here, we obtained the annual number of newly-diagnosed AIP cases, age, gender, and consultation departments from January 2011 to December 2020 through retrieving the medical records of inpatients from 32 comprehensive grade III A hospitals in Hebei, China. Based on the principle of confidentiality, patient names, contact information, and medical histories were not involved. The population data of Hebei province were obtained from Hebei Provincial Bureau of Statistics (tjj.hebei.gov.cn/). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of the Second Hospital of Hebei Medical University (2021-R228). Informed consent was waived by the Ethics Committee as this is a retrospective study. These 32 hospitals were also informed and approved the study.
Statistical analysis
Confidence intervals were calculated by VassarStats (http://vassarstats.net/index.html). The software SPSS 25.0 (IBM Corp., Armonk, NY, USA) was used for statistical analysis. The normal distribution test was utilized to estimate how to describe the data. Normally distributed continuous data were represented by mean ± standard deviation (). Non-normally distributed data were expressed as the median (P25, P75).
Results
Prevalence and characteristics of pathogenic HMBS gene variants in Chinese and other racial groups
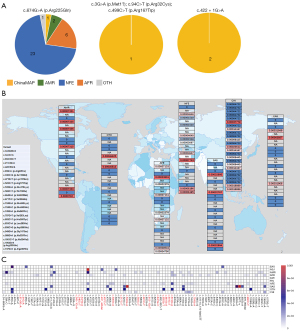
A total of 501 HMBS gene variants were detected in ChinaMAP. According to the ACMG guidelines, 5 P/LP variants were identified including c.3G>A (p. Met1?), c.94C>T (p. Arg32Cys), c.422+1G>A, c.499C>T (p. Arg167Trp), and c.674G>A (p. Arg225Gln). The prevalence of pathogenic HMBS variants in the Chinese population was estimated to be 1/1,765. The 5 variants were all recoded in the Human Gene Mutation Database (HGMD; http://www.hgmd.cf.ac.uk/ac/index.php) as “diseases-causing mutation (DM)”, and c.94C>T (p. Arg32Cys), c.499C>T (p. Arg167Trp) and c.674G>A (p. Arg225Gln) were located in the CpG dinucleotide, the mutation hotspot (Table 1, Figure 1A). In addition, 25 VUS-P variants were identified (Figure 1A). Variation types of all P/LP and VUS-P variants in ChinaMAP are summarized in Figure 1B, among which nonsynonymous variant was the most common type.
Table 1
| c.DNA change | Protein change | CpG dinucleotide | Variant type | Reported in HGMD | Variant assessment by ACMG | Evidence of pathogenicity | REVELa | s-PP3b | Allele count | Allele frequency | prevalence of pathogenic HMBS variants in Chinese |
|---|---|---|---|---|---|---|---|---|---|---|---|
| c.3G>A | p. Met1? | − | Start-loss | DM | LP | PVS1_Moderate, PM2_supporting, PS3_Moderate, PS4_supporting | 0.696 | − | 1 | 0.000047 | 1/5,294 |
| c.94C>T | p. Arg32Cys | + | Nonsynonymous variant | DM | LP | PM2_supporting, PP3; PM5; PM1 | 0.852 | − | 1 | 0.000047 | 1/5,294 |
| c.422+1G>A | − | − | Canonical splicing | DM | LP | PVS1, PM2_supporting | − | 3 [ADA, RF, spliceAI] | 2 | 0.000094 | 1/10,588 |
| c.499C>T | p. Arg167Trp | + | Nonsynonymous variant | DM | P | PM2_supporting, PP3; PM5; PS3, PS4_Moderate | 0.943 | − | 1 | 0.000047 | 1/5,294 |
| c.674G>A | p. Arg225Gln | + | Nonsynonymous variant | DM | LP | PM2_supporting, PP3; PM1; PM5; PS4_supporting: | 0.915 | − | 1 | 0.000047 | 1/5,294 |
| Prevalence of total pathogenic HMBS variants in Chinese | 1/1,765 | ||||||||||
DNA and protein reference sequences are RefSeq NM_000190.4 and NP_000181.2, respectively. aREVEL: a score that combines the results of multiple software predictions including SIFT and Polyphen etc. PP3 evidence of ACMG guideline can be used when REVEL score >0.75. bs-PP3: the score of five splicing prediction software (dbscSNV_ADA, dbscSNV_RF, MMsplice, MaxEnt, SpliceAI) are synthesized. Score 1 point for each piece of software that is deemed to affect splicing. +, the variant is located in CpG dinucleotide; −, the variant is not located in CpG dinucleotide. DM, diseases-causing mutation; P, pathogenic; LP, likely pathogenic.
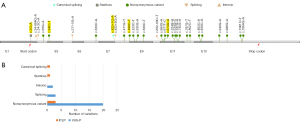
Based on the ACMG guidelines, 20 P/LP and 74 VUS-P variants were screened out from the 2,314 variants of HMBS identified in GnomAD Genome V3.0 (Table 2, Figure 2A,2B). Nonsynonymous variants appeared most frequently (Figure 2C). The predicted prevalence of pathogenic HMBS variants was 1/1,367, 1/1,403, and 1/621 in AMR, AFR, and NFE, respectively (Table 3).
Table 2
| c.DNA change | Protein change | Variant assessment by ACMG | Variant type | Evidence of pathogenicity | Reported in HGMD | REVELa | s-PP3b |
|---|---|---|---|---|---|---|---|
| c.674G>A | p. Arg225Gln | LP | Nonsynonymous variant | PM2_supporting, PP3; PM1; PM5; PS4_supporting | DM | 0.915 | – |
| c.91G>A | p. Ala31Thr | LP | Nonsynonymous variant | PM2_supporting, PM5, PP3, PM1, PS4_supporting | DM | 0.985 | – |
| c.345-1G>A | – | LP | Canonical splicing | PVS1, PM2_supporting | DM | - | 5[ADA, RF, MMsplice, maxent, spliceAI] |
| c.346C>T | p. Arg116Trp | P | Nonsynonymous variant | PM2_supporting, PM5, PP3, PM1, PS4 | DM | 0.95 | – |
| c.347G>A | p. Arg116Gln | LP | Nonsynonymous variant | PM2_supporting, PM5, PP3, PM1 | DM | 0.953 | – |
| c.457C>T | p. Gln153* | LP | Stopgain | PVS1, PM2_supporting | – | – | 0.5[spliceAI] |
| c.500G>A | p. Arg167Gln | P | Nonsynonymous variant | PM2_supporting, PP3; PM5, PS3; PS4_Moderate | DM | 0.934 | – |
| c.517C>T | p. Arg173Trp | P | Nonsynonymous variant | PM2_supporting, PP3; PM5, PM1; PS4 | DM | 0.903 | – |
| c.532G>A | p. Asp178Asn | LP | Nonsynonymous variant | PM2_supporting, PP3; PM5, PM1; PS4_supporting | DM | 0.861 | – |
| c.583C>T | p. Arg195Cys | P | Nonsynonymous variant | PM2_supporting, PP3; PM5, PM1; PS3 | DM | 0.903 | – |
| c.601C>T | p. Arg201Trp | P | Nonsynonymous variant | PM2_supporting, PP3; PM5, PM1; PS3; PS4_Moderate | DM | 0.925 | – |
| c.613-1G>T | – | P | Canonical splicing | PVS1, PM2_supporting; PP1_Moderate | DM | – | 5[ADA, RF, MMsplice, maxent, spliceAI] |
| c.655G>T | p. Ala219Ser | LP | Nonsynonymous variant | PM2_supporting, PP3, PM1, PM5 | – | 0.873 | – |
| c.673C>T | p. Arg225* | P | Stopgain | PVS1, PM2_supporting; PS3, PS4_supporting | DM | – | 1.5[maxent, spliceAI] |
| c.754G>A | p. Ala252Thr | LP | Nonsynonymous variant | PM2_supporting, PP3; PM5, PM1 | DM | 0.793 | – |
| c.992C>T | p. Ala331Val | LP | Nonsynonymous variant | PM2_supporting, PP3, PM1, PS3 | DM | 0.962 | – |
| c.661G>A | p. Gly221Ser | LP | Nonsynonymous variant | PM2_supporting, PP3; PM5; PM1 | – | 0.798 | – |
| c.87+5G>A | – | LP | Splicing | PM2_supporting, PP3; PS3 | DM | – | 5[ADA, RF, MMsplice, maxent, spliceAI] |
| c.437_443del | p. Ser147Glufs*106 | LP | Frameshift deletion | PVS1, PM2_supporting | DM | – | – |
| c.647G>C | p. Gly216Ala | LP | Nonsynonymous variant | PM2_supporting, PP3; PM1; PM5 | DM | 0.866 |
DNA and protein reference sequences are RefSeq NM_000190.4 and NP_000181.2, respectively. aREVEL: a score that combines the results of multiple software predictions including SIFT and Polyphen etc. PP3 evidence of ACMG guideline can be used when REVEL score >0.75. bs-PP3: the score of five splicing prediction software (dbscSNV_ADA, dbscSNV_RF, MMsplice, MaxEnt, SpliceAI) are synthesized. Score 1 point for each piece of software that is deemed to affect splicing. P, pathogenic; LP, likely pathogenic; DM, diseases-causing mutation; ACMG, American College of Medical Genetics and Genomics.
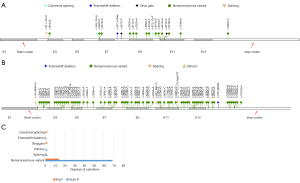
Table 3
| c.DNA change | Protein change | NFE | AMR | AFR | SAS | ASJ | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele count | Allele frequency | Allele count | Allele frequency | Allele count | Allele frequency | Allele count | Allele frequency | Allele count | Allele frequency | ||||||
| c.674G>A | p. Arg225Gln | 23 | 0.000356258 | 2 | 0.000146477 | 6 | 0.000142742 | 0 | 0 | 0 | 0 | ||||
| c.91G>A | p. Ala31Thr | 1 | 0.0000154842 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| c.345-1G>A | – | 0 | 0 | 1 | 0.0000732493 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| c.346C>T | p. Arg116Trp | 0 | 0 | 0 | 0 | 1 | 0.0000237903 | 0 | 0 | 0 | 0 | ||||
| c.347G>A | p. Arg116Gln | 2 | 0.0000309684 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| c.457C>T | p. Gln153* | 0 | 0 | 1 | 0.0000732172 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| c.500G>A | p. Arg167Gln | 7 | 0.000108413 | 0 | 0 | 2 | 0.0000475489 | 0 | 0 | 0 | 0 | ||||
| c.517C>T | p. Arg173Trp | 1 | 0.0000154856 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| c.532G>A | p. Asp178Asn | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0.00131234 | 0 | 0 | ||||
| c.583C>T | p. Arg195Cys | 1 | 0.0000154832 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| c.601C>T | p. Arg201Trp | 4 | 0.0000619464 | 0 | 0 | 1 | 0.0000237722 | 0 | 0 | 0 | 0 | ||||
| c.613-1G>T | – | 0 | 0 | 0 | 0 | 1 | 0.0000237891 | 0 | 0 | 0 | 0 | ||||
| c.655G>T | p. Ala219Ser | 5 | 0.0000774305 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| c.673C>T | p. Arg225* | 1 | 0.0000154923 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| c.754G>A | p. Ala252Thr | 2 | 0.0000309684 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| c.992C>T | p. Ala331Val | 4 | 0.0000619598 | 0 | 0 | 2 | 0.0000475805 | 0 | 0 | 0 | 0 | ||||
| c.661G>A | p. Gly221Ser | 0 | 0 | 0 | 0 | 1 | 0.0000237869 | 0 | 0 | 0 | 0 | ||||
| c.87+5G>A | – | 1 | 0.0000154847 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| c.437_443del | p. Ser147Glufs*106 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0.000300842 | ||||
| c.647G>C | p. Gly216Ala | 0 | 0 | 1 | 0.0000732601 | 1 | 0.0000237993 | 0 | 0 | 0 | 0 | ||||
| Prevalence of pathogenic HMBS variants | 1/621 | 1/1,367 | 1/1,403 | 1/382a | 1/1,662b | ||||||||||
DNA and protein reference sequences are RefSeq NM_000190.4 and NP_000181.2, respectively. aSAS/bASJ: the results may be inaccurate because of the too small number of total alleles. ASJ, Ashkenazi Jewish; AMR, Mixed American; AFR, African/African American; NFE, Non-Finnish European; SAS, South Asian.
Noticeably, some variants listed as DM in HGMD were not considered P/LP according to the ACMG guidelines, such as c.569C>T (p. Thr190Ile) and c.664G>A (p. Val222Met), which were interpreted as VUS-P.
Distribution characteristics of P/LP and VUS-P HMBS variants among various racial groups
As demonstrated in Table 3 and Figure 3A-3C, the distribution and prevalence of these P/LP and VUS-P variants varied among different racial groups. For example, c.674G>A (p. Arg225Gln) was the most widely distributed variant among all P/LP variants identified in both ChinaMAP and GnomAD Genome V3.0, which presented in group CHI, AMR, AFR, and NFE. The remaining four P/LP variants identified in ChinaMAP [c.3G>A (p. Met1?); c.94C>T (p. Arg32Cys); c.422+1G>A; c.499C>T (p. Arg167Trp] were not available in the populations in GnomAD Genome V3.0. In addition, c.532G>A (p. Asp178Asn) was detected only in SAS, with a high allele frequency 0.001312 (4/3,048). Furthermore, c.457C>T (p. Gln153*) presented only in AMR, whereas c.613-1G>T and c.661G>A only in AFR. The number of pathogenic variant sites identified in NFE was the largest, including 12 sites (Table 3).
Epidemiological survey of AIP in Hebei Province, China
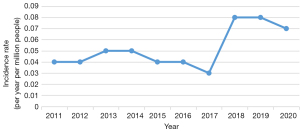
The annual incidence of AIP in Hebei province between 2011 and 2020 was estimated to be from 0.03 [95% confidence interval (CI): 0.01 to 0.11] to 0.08 (95% CI: 0.03 to 0.18) per million inhabitants. It was generally low between 2011 and 2017, but increased in 2018 and became roughly stable thereafter (Table 4, Figure 4). A total of 39 new cases including 36 females and 3 males were diagnosed with AIP during these 10 years. The age at diagnosis ranged from 15 to 46 years old, presenting a normal distribution, and 25.59±6.66 years old on average. The patients chose to consult multiple departments, such as Department of Endocrinology, Department of Hematology, Department of Neurology, and Department of Psychiatry, as illustrated in Table 4.
Table 4
| Year | Number of new patients | Population (millions) | Incidence (per million population) | Gender | Department of patient visited |
|---|---|---|---|---|---|
| 2011 | 3 | 72.41 | 0.04 (0.01–0.13) | F | Hematology [1]; Gastroenterology [1]; Surgery [1] |
| 2012 | 3 | 72.88 | 0.04 (0.01–0.13) | F | Gastroenterology [2]; Neurology [1] |
| 2013 | 4 | 73.33 | 0.05 (0.02–0.14) | F | Neurology [1]; Hematology [1]; Gastroenterology [1]; Gynecology [1] |
| 2014 | 4 | 73.84 | 0.05 (0.02–0.14) | F | Gastroenterology [3]; Surgery [1] |
| 2015 | 3 | 74.25 | 0.04 (0.01–0.13) | F | Gastroenterology [2]; Surgery [1] |
| 2016 | 3 | 74.70 | 0.04 (0.01–0.13) | F2; M1 | Gastroenterology [2]; Emergency Department [1] |
| 2017 | 2 | 75.20 | 0.03 (0.01–0.11) | F | Hematology [1]; Neurology [1] |
| 2018 | 6 | 75.56 | 0.08 (0.03–0.18) | F4; M2 | Gastroenterology [2]; Rehabilitation [1]; Emergency Department [1]; General Practice [1]; Endocrinology [1] |
| 2019 | 6 | 75.92 | 0.08 (0.03–0.18) | F | Endocrinology [3]; Psychiatry [1]; General Practice [1]; Gastroenterology [1] |
| 2020 | 5 | 74.61 | 0.07 (0.03–0.17) | F | Endocrinology [3]; Hematology [1]; Gastroenterology [1] |
Figures in parentheses are 95% confidence limits. AIP, acute intermittent porphyria; M, male; F, female.
Discussion
To date, over 500 different variants in HMBS gene have been listed in HGMD. In this study, the prevalence of pathogenic HMBS variants in the Chinese population was estimated and the epidemiological features of AIP patients admitted to tertiary hospitals in Hebei Province, China were investigated.
In the WGS database ChinaMAP, we identified 5 pathogenic HMBS variants and the prevalence of pathogenic HMBS variants in the general Chinese population was estimated to be 1/1,765, suggesting that the number of people with pathogenic HMBS variants may exceed 790,000 in China, a country with 1.4 billion population. Although the penetrance of AIP is very low, all individuals with pathogenic HMBS variants are at risk of developing acute attacks. Furthermore, findings have demonstrated that chronic complications, such as hepatocellular carcinoma, hypertension, and chronic renal failure could occur in not only symptomatic AIP, but also latent patients with pathogenic HMBS variants. Many triggers have been known to cause acute episodes in carriers of pathogenic HMBS variants, while early standardized management and avoidance of triggers can reduce acute attacks in these potential patients. Thus, these individuals with pathogenic HMBS variants in China should be given high priority for further preventive strategy.
Next, the distribution characteristics of pathogenic HMBS variants were described among various racial populations. From the GnomAD Genome V3.0 database, 20 pathogenic HMBS variants were identified and the prevalence of pathogenic HMBS variants in some different racial populations was predicted as follows: 1/1,367 in AMR, 1/1,403 in AFR, and 1/621 in NFE. In line with previous findings (7), nonsynonymous variant was the most frequent of all P/LP and VUS-P variants in databases of our study. Studies have explored whether AIP has significant molecular genetic heterogeneity, and only a few variants have relatively commonly been attributed to either occur in CpG dinucleotide or due to founder effect, such as HMBS p. Trp198* in Sweden (13,16). Among all P/LP HMBS variants in our project, c.674G>A (p. Arg225Gln) was widely identified in AMR, AFR, NFE, and CHI populations, which may be related to its occurrence in CpG dinucleotide. The CpG dinucleotides, known as the mutation hot spots, are confirmed to be hypermutable because of oxidative deamination of methylated cytosines (17). The distribution and detected frequency of different variants varied in various racial groups. For instance, c.532G>A (p. Asp178Asn) and c.457C>T (p. Gln153*) was recorded only in SAS and AMR, respectively. However, c.613-1G>T and c.661G>A presented only in AFR. Consequently, the remarkable genetic heterogeneity of AIP was revealed, indicating that it is necessary to understand their respective genetic characteristics to manage different racial patients effectively. Furthermore, the prevalence of pathogenic HMBS variants in NFE was as high as 1/621 with 12 identified sites. The result was bound up with the high prevalence of AIP in Europe, particularly in countries such as Sweden, where the founder effect exists.
In an early report, the predicted prevalence of healthy French blood donors with reduced HMBS enzyme activity was 1/1,675 (18). Nevertheless, 2 isoforms of HMBS exist, the housekeeping and the erythroid isoforms. The transcript including exons 1 and 3–15 is directed by housekeeping promoter and the transcript containing exons 2–15 is produced by an erythroid-specific promoter (19,20). When variants occur within or adjacent to the coding region of exon 1, the activity of HMBS in erythroid cells is normal (21-23). Moreover, erythrocyte enzyme activity is tied up with multiple factors, such as erythrocyte status and personal health (24). Anemia, liver disease, and some genetic factors may result in the increase or decrease of HMBS activity in red blood cells (25). Consequently, the scheme may underestimate the prevalence.
Thanks to rapid development of next-generation sequencing (NGS) technology, gene sequencing has been widely applied in AIP diagnosis in recent years. Using open database information, 2 studies estimated the prevalence of pathogenic HMBS in a predominantly Caucasian population of 1/1,782 and 1/1,299 (6,7). The pathogenicity of HMBS variants in both studies was assessed according to the functional in vitro expression and thermostability studies, combined with in silico analyses. Results about variant c.674G>A (p. Arg225Gln) in the 2 studies piqued our attention. It was considered as “pathogenic” in silico analyses in both studies, but inconsistent results appeared after in vitro functional verification. The activity of HMBS was (102±19)% of wild-type (WT) activity, with no abnormal thermal stability in 1 study (6). However, in another study, the enzyme activity was (65±3.5)% of WT activity with poor thermostability, and it was proposed that if the variant was considered pathogenic, the prevalence of the pathogenic HMBS variants can be increased to 1/650 (7), which was consistent with our prediction of 1/621 in NFE. The International Porphyrin Collaboration has been working to identify pathogenic porphyrin gene variants (26) and currently recommends the ACMG guidelines as the criteria for interpreting variants. On the basis of ACMG guidelines, we found that c.674G>A (p. Arg225Gln) was detected with extremely low frequency (<0.05%) in publicly available population databases, and occurred in mutational hotspot. In addition, the missense variant in the same position, such as c.673C>G (p.Arg225Gly) has been shown to be pathogenic (27,28). Moreover, it was predicted as “pathogenic” by many algorithms and 3 patients with this variant have been reported (16). Thus, the variant was interpreted as LP. Although functional expression in vitro and thermal stability study can predict the pathogenicity of variants better than in silico analysis, there are still certain limitations. Previous studies have revealed that the residual activity of HMBS enzyme expressed in vitro is not consistent with the activity in red blood cells in vivo (29), and the enzyme activity in red blood cells failed to reflect the level in liver accurately (30). Thus, functional studies should be utilized cautiously and its reproducibility should be focused on (31). In addition, some variants listed as pathogenic in HGMD, such as c.569C>T (p. Thr190Ile) and c.664G>A (p. Val222Met), were classified as VUS-P on the basis of the ACMG guidelines, not P or LP. In summary, it is necessary to evaluate the pathogenicity of variants according to the ACMG guidelines and criteria.
Our study has some limitations. In the present study, we also identified a number of VUS-P variants (25 in ChinaMAP; 74 in GnomAD genome V3.0), some of which may be confirmed as P/LP with more clinical cases and functional verification, suggesting that our current prediction of prevalence of pathogenic HMBS variants should be “minimal”.
To understand the incidence and other epidemiological characteristics of Chinese patients with AIP, an epidemiological investigation of AIP in China needs to be carried out, and we performed it in Hebei Province as a start. Here, we conducted a retrospective study in Hebei, which lies in the north of China with a population of over 70 million. Combined with the prevalence of pathogenic HMBS variants of 1/1,765 and a penetrance of 1%, it is estimated that there should be approximately 400 symptomatic AIP patients here. Nevertheless, only 39 AIP patients have been diagnosed over 10 years, indicating a large proportion of AIP patients remain undiagnosed.
We found the apparent inconstancy of annual incidence of AIP in Hebei province over 10 years, ranging from 0.03 (95% CI: 0.01 to 0.11) to 0.05 (95% CI: 0.07 to 0.14) per million population between 2011 and 2017. Nevertheless, it amounted to 0.08 (95% CI: 0.03 to 0.18) per million inhabitants in 2018, generally stabilizing thereafter.
This fluctuation of annual incidence may be related to the following factors. The misdiagnosis rate of AIP in China is very high in early years due to poor awareness of this rare disease among Chinese clinicians (32). Clinical phenotypic heterogeneity and non-specificity are critical clinical features of AIP (33), mirrored in our investigation by the multiple departments visited by AIP patients. It is a neurovisceral disease caused by the accumulation of PBG and ALA, which leads to multisystemic damage (34,35). Abdominal pain is the most common manifestation of patients with AIP; the symptom of pain is typically disproportionate to the exam findings on physical examination (36). Therefore, gastroenterology, general surgery, and even gynecology may be the departments where AIP patients visit for the first time or repeatedly. Both the peripheral and central nervous system can be damaged due to the factors, such as neurotoxicity, of ALA. It can also cause the syndrome of improper vasopressin secretion (SIDAI), leading to severe hyponatremia (4). These are probably the reasons for AIP patients to consult the Departments of Neurology, Rehabilitation, and Endocrinology. Furthermore, psychiatric symptoms have been confirmed as occurring in patients with AIP in the form of depression, anxiety, hallucinations, psychosis, or delirium (37). In our study, 1 case was diagnosed in the psychiatric department. Besides, hypertension, tachycardia, and renal dysfunction are also frequent clinical manifestations. Consequently, clinicians in various departments should have the capability to make an accurate diagnosis of AIP.
The accurate diagnosis of AIP is limited by the lack of some essential tests. During the survey, some cases were diagnosed as “porphyria” and “porphyria?”. Urinary ALA and plasma emission peak play the key roles in the diagnosis and typing of porphyria, whereas few hospitals in China can conduct these tests. Recently, gene detection has been widely used in the diagnosis and differential diagnosis of AIP. Medical professionals in China have begun to realize the importance of genetic testing for accurate diagnosis (38).
A series of initiatives have been carried out in recent years, attracting increasing attention from the Chinese government to rare diseases. From the beginning of 2016, National Health and Family Planning Commission began to build the Rare Disease Diagnosis and Treatment and Protection Expert Committee. In early 2018, 5 departments including National Health Commission (NHC) jointly formulated the First Batch of Rare Diseases Catalogue, and then the China Alliance for Rare Diseases (CARD; https://www.chard.org.cn/) was established. Consequently, this rare disorder has garnered attention from Chinese clinicians, which is the most likely the reason for the apparent increase of AIP incidence in 2018 and thereafter in our research.
A study of 11 European countries indicated that the annual incidence of symptomatic AIP was 0.13 per million population and the prevalence was 5.9 per million population. In addition, the numbers were similar in all countries except Sweden. Prevalence rates were significantly high in some regions due to the founder effect, such as 23 per million in Sweden and 17.7 per million in south-eastern Spain (13). Notably, the incidence of symptomatic AIP in Europe has been sliding, which was attributed to the factors that Europe paid attention to porphyria earlier, established a perfect diagnosis and management system for patients with AIP, and strengthened family screening and prevention consultation (30).
As an autosomal dominant disease, no gender differences in gene carriers are expected in AIP patients. Nevertheless, all studies have indicated that acute attacks affect females more frequently than males (9,13,39,40). A majority of patients were aged from 20 to 40 years old upon onset, with few cases of before adolescence and after menopause (9,13,40). Similarly, the number of female patients was significantly larger than that of their male counterparts in our survey and the mean age was 25.59±6.66 (ranging from 15 to 46) years, suggesting that female hormones probably play a role in the onset of AIP. We know that several precipitants of AIP, including increased progesterone, a common female hormone, can trigger AIP by inducing ALAS production (30,41).
Our study has some limitations. Due to regional differences, the epidemiological characteristics of AIP in Hebei province cannot fully represent that of the whole China. However, it helps us to understand the status of AIP in local areas of China, and emphasizes the necessity of carrying out a nationwide epidemiological investigation of AIP.
Conclusions
Taken together, approximately 400 symptomatic AIP patients are estimated to exist in Hebei, whereas only 39 AIP patients have been diagnosed over 10 years, indicating a large proportion of AIP patients remain unidentified. Currently, China has made great strides in the management of rare diseases including AIP. More nationwide epidemiological surveys and studies related to the penetrance rate of AIP patients in China should be performed.
Acknowledgments
Funding: This study was supported by Grants from Hebei Provincial Science and Technology Plan Project (21377739D).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1600/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1600/dss
Conflicts of Interest: Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1600/coif). All authors reported that this study was supported by Grants from Hebei Provincial Science and Technology Plan Project (21377739D). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of the Second Hospital of Hebei Medical University (2021-R228).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Puy H, Gouya L, Deybach JC. Porphyrias. Lancet 2010;375:924-37. [Crossref] [PubMed]
- Wang B, Rudnick S, Cengia B, et al. Acute Hepatic Porphyrias: Review and Recent Progress. Hepatol Commun 2018;3:193-206. [Crossref] [PubMed]
- Bung N, Roy A, Chen B, et al. Human hydroxymethylbilane synthase: Molecular dynamics of the pyrrole chain elongation identifies step-specific residues that cause AIP. Proc Natl Acad Sci U S A 2018;115:E4071-80. [Crossref] [PubMed]
- Spiritos Z, Salvador S, Mosquera D, et al. Acute Intermittent Porphyria: Current Perspectives And Case Presentation. Ther Clin Risk Manag 2019;15:1443-51. [Crossref] [PubMed]
- Naik H, Stoecker M, Sanderson SC, et al. Experiences and concerns of patients with recurrent attacks of acute hepatic porphyria: A qualitative study. Mol Genet Metab 2016;119:278-83. [Crossref] [PubMed]
- Chen B, Solis-Villa C, Hakenberg J, et al. Acute Intermittent Porphyria: Predicted Pathogenicity of HMBS Variants Indicates Extremely Low Penetrance of the Autosomal Dominant Disease. Hum Mutat 2016;37:1215-22. [Crossref] [PubMed]
- Lenglet H, Schmitt C, Grange T, et al. From a dominant to an oligogenic model of inheritance with environmental modifiers in acute intermittent porphyria. Hum Mol Genet 2018;27:1164-73. [Crossref] [PubMed]
- Manceau H, Gouya L, Puy H. Acute hepatic and erythropoietic porphyrias: from ALA synthases 1 and 2 to new molecular bases and treatments. Curr Opin Hematol 2017;24:198-207. [Crossref] [PubMed]
- Bonkovsky HL, Maddukuri VC, Yazici C, et al. Acute porphyrias in the USA: features of 108 subjects from porphyrias consortium. Am J Med 2014;127:1233-41. [Crossref] [PubMed]
- Lin CN, Shiao MS, Cheng ML, et al. Profiling of Serum Metabolites of Acute Intermittent Porphyria and Asymptomatic HMBS Mutation Carriers. Cells 2021;10:2579. [Crossref] [PubMed]
- Buendía-Martínez J, Barreda-Sánchez M, Rodríguez-Peña L, et al. Health impact of acute intermittent porphyria in latent and non-recurrent attacks patients. Orphanet J Rare Dis 2021;16:106. [Crossref] [PubMed]
- Kauppinen R, Mustajoki P. Prognosis of acute porphyria: occurrence of acute attacks, precipitating factors, and associated diseases. Medicine (Baltimore) 1992;71:1-13. [Crossref] [PubMed]
- Elder G, Harper P, Badminton M, et al. The incidence of inherited porphyrias in Europe. J Inherit Metab Dis 2013;36:849-57. [Crossref] [PubMed]
- Gilles A, Vermeersch S, Vermeersch P, et al. Expert consensus statement on acute hepatic porphyria in Belgium. Acta Clin Belg 2021; [Epub ahead of print]. [Crossref] [PubMed]
- Cao Y, Li L, Xu M, et al. The ChinaMAP analytics of deep whole genome sequences in 10,588 individuals. Cell Res 2020;30:717-31. [Crossref] [PubMed]
- Floderus Y, Shoolingin-Jordan PM, Harper P. Acute intermittent porphyria in Sweden. Molecular, functional and clinical consequences of some new mutations found in the porphobilinogen deaminase gene. Clin Genet 2002;62:288-97. [Crossref] [PubMed]
- Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet 1988;78:151-5. [Crossref] [PubMed]
- Nordmann Y, Puy H, Da Silva V, et al. Acute intermittent porphyria: prevalence of mutations in the porphobilinogen deaminase gene in blood donors in France. J Intern Med 1997;242:213-7. [Crossref] [PubMed]
- Chretien S, Dubart A, Beaupain D, et al. Alternative transcription and splicing of the human porphobilinogen deaminase gene result either in tissue-specific or in housekeeping expression. Proc Natl Acad Sci U S A 1988;85:6-10. [Crossref] [PubMed]
- Yoo HW, Warner CA, Chen CH, et al. Hydroxymethylbilane synthase: complete genomic sequence and amplifiable polymorphisms in the human gene. Genomics 1993;15:21-9. [Crossref] [PubMed]
- Chen CH, Astrin KH, Lee G, et al. Acute intermittent porphyria: identification and expression of exonic mutations in the hydroxymethylbilane synthase gene. An initiation codon missense mutation in the housekeeping transcript causes "variant acute intermittent porphyria" with normal expression of the erythroid-specific enzyme. J Clin Invest 1994;94:1927-37. [Crossref] [PubMed]
- Grandchamp B, Picat C, Kauppinen R, et al. Molecular analysis of acute intermittent porphyria in a Finnish family with normal erythrocyte porphobilinogen deaminase. Eur J Clin Invest 1989;19:415-8. [Crossref] [PubMed]
- Granata F, Mendez M, Brancaleoni V, et al. Molecular characterization, by digital PCR analysis of four HMBS gene mutations affecting the ubiquitous isoform of Porphobilinogen Deaminase (PBGD) in patients with Acute Intermittent Porphyria (AIP). Mol Genet Metab 2018;125:295-301. [Crossref] [PubMed]
- Grandchamp B, Nordmann Y. Enzymes of the heme biosynthesis pathway: recent advances in molecular genetics. Semin Hematol 1988;25:303-11. [PubMed]
- Mustajoki P, Kauppinen R, Lannfelt L, et al. Frequency of low erythrocyte porphobilinogen deaminase activity in Finland. J Intern Med 1992;231:389-95. [Crossref] [PubMed]
- Chen B, Whatley S, Badminton MInternational Porphyria Molecular Diagnostic Collaborative, et al. an evidence-based database of verified pathogenic and benign variants for the porphyrias. Genet Med 2019;21:2605-13. [Crossref] [PubMed]
- Kauppinen R, Mustajoki S, Pihlaja H, et al. Acute intermittent porphyria in Finland: 19 mutations in the porphobilinogen deaminase gene. Hum Mol Genet 1995;4:215-22. [Crossref] [PubMed]
- Mustajoki S, Laine M, Lahtela M, et al. Acute intermittent porphyria: expression of mutant and wild-type porphobilinogen deaminase in COS-1 cells. Mol Med 2000;6:670-9. [Crossref] [PubMed]
- Ulbrichova D, Schneider-Yin X, Mamet R, et al. Correlation between biochemical findings, structural and enzymatic abnormalities in mutated HMBS identified in six Israeli families with acute intermittent porphyria. Blood Cells Mol Dis 2009;42:167-73. [Crossref] [PubMed]
- Fraunberg MVUZ, Pischik E, Udd L, et al. Clinical and biochemical characteristics and genotype-phenotype correlation in 143 Finnish and Russian patients with acute intermittent porphyria. Medicine (Baltimore) 2005;84:35-47. [Crossref] [PubMed]
- Brnich SE, Abou Tayoun AN, Couch FJ, et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med 2019;12:3. [Crossref] [PubMed]
- Ren Y, Xu LX, Liu YF, et al. A novel 55-basepair deletion of hydroxymethylbilane synthase gene found in a Chinese patient with acute intermittent porphyria and her family: A case report. Medicine (Baltimore) 2018;97:e12295. [Crossref] [PubMed]
- Balwani M, Wang B, Anderson KE, et al. Acute hepatic porphyrias: Recommendations for evaluation and long-term management. Hepatology 2017;66:1314-22. [Crossref] [PubMed]
- Floderus Y, Sardh E, Möller C, et al. Variations in porphobilinogen and 5-aminolevulinic acid concentrations in plasma and urine from asymptomatic carriers of the acute intermittent porphyria gene with increased porphyrin precursor excretion. Clin Chem 2006;52:701-7. [Crossref] [PubMed]
- Gerischer LM, Scheibe F, Nümann A, et al. Acute porphyrias - A neurological perspective. Brain Behav 2021;11:e2389. [Crossref] [PubMed]
- Balwani M, Desnick RJ. The porphyrias: advances in diagnosis and treatment. Hematology Am Soc Hematol Educ Program 2012;2012:19-27. [Crossref] [PubMed]
- Duque-Serrano L, Patarroyo-Rodriguez L, Gotlib D, et al. Psychiatric Aspects of Acute Porphyria: a Comprehensive Review. Curr Psychiatry Rep 2018;20:5. [Crossref] [PubMed]
- Hu Y, Li W, Kang N, et al. Identification and molecular analysis of 17 novel variants of hydroxymethylbilane synthase in Chinese patients with acute intermittent porphyria. Clin Genet 2022;101:116-21. [Crossref] [PubMed]
- Baumann K, Kauppinen R. Penetrance and predictive value of genetic screening in acute porphyria. Mol Genet Metab 2020;130:87-99. [Crossref] [PubMed]
- Schuurmans MM, Schneider-Yin X, Rüfenacht UB, et al. Influence of age and gender on the clinical expression of acute intermittent porphyria based on molecular study of porphobilinogen deaminase gene among Swiss patients. Mol Med 2001;7:535-42. [Crossref] [PubMed]
- Bonkovsky HL, Dixon N, Rudnick S. Pathogenesis and clinical features of the acute hepatic porphyrias (AHPs). Mol Genet Metab 2019;128:213-8. [Crossref] [PubMed]
(English Language Editor: J. Jones)


