
Network pharmacology and bioinformatics analysis identified essential genes of Jingulian in the treatment of rheumatoid arthritis and COVID-19
Introduction
Coronavirus disease-19 (COVID-19) has been a global pandemic disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a new coronavirus discovered in 2019 (1). Remdesivir was the only drug approved by the U.S. Food and Drug Administration (FDA) to treat COVID-19 in adults and children aged 12 and older (2). However, this drug shows limited therapeutic effectiveness especially in patients with severe COVID-19. Therefore, more effective drugs need to be discovered to treat patients with complicated COVID-19.
Rheumatoid arthritis (RA) is a chronic inflammatory joint disease characterized by synovial hyperproliferation and inflammatory cell infiltration, which can lead to progressive joint damage and even disability, seriously affecting the quality of life of patients (3,4). Furthermore, in-hospital patients with RA are more likely to be infected with COVID-19. In addition, patients with RA are more likely to have a higher risk of severe COVID-19 outcomes (5). Meanwhile, there is also some evidence that COVID-19 may increase the risk of developing RA (6). Treatment for patients with COVID-19 and RA is difficult due to a dysfunction of the immune system and lack of effective medicines. Hence, effective treatments should be explored in such patients.
Jingulian, a traditional Chinese medicine (TCM), can obviously dampen inflammatory responses and provides symptom relief for patients with RA. Jingulian consists of 5 TCMs, including Alangium (Alangiaceae; Chinese alangium), Sargentodoxa Caulis (Lardizabalaceae; Sargentodoxa cuneata), Speranskiae seulmpaticntis Herba (Ericaceae; Gaultheria trichophylla Royle), Psammosilenetunicoides (Caryophyllaceae; Psammosilene), and Guangxi schefflera twig and leaf (Araliaceae; Schefflera leucantha R. Viguier). However, the underlying mechanisms of Jingulian in the treatment of patients with RA/COVID-19 remain unknown.
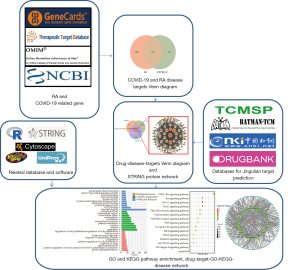
Network pharmacology is a promising method used to explore all candidate targets and underlying mechanisms of a particular disease (7). Molecular docking is a useful bioinformatics tool used to explore the behaviors of ingredients at the binding site of a target protein (8). In our study, network pharmacology analysis and molecular docking were employed to evaluate the pharmacological and detailed mechanisms of Jingulian. The workflow is shown in Figure 1. This study reveals the potential targets and pathways of active compounds derived from Jingulian against patients with RA and COVID-19. Moreover, this study provides new insights into the detailed mechanisms of traditional Chinese medicine against RA and COVID-19. We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1665/rc).
Methods
Schematic diagram
The schematic diagram of the research methods in our study is shown in Figure 1. Network pharmacology approaches and molecular docking were employed to explore the pharmacological and detailed mechanisms of Jingulian in the treatment of RA and COVID-19. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of the active chemical components
With the purpose of retrieving the chemical ingredients of components in Jingulian capsule, we conducted a complete search using the Traditional Chinese Medicine Systems Pharmacology database (TCMSP, https://old.tcmsp-e.com/tcmsp.php) (9), traditional Chinese medicine database@TAIWAN database (TCM@TAIWAN, http://tcm.cmu.edu.tw/zh-tw/) (10), and China National Knowledge Infrastructure (CNKI, https://www.cnki.net). The names of herbs were used as the main words to acquire all components. Based on absorption, distribution, metabolism, and excretion, active compounds of Jingulian were screened. In screening out the compounds, oral bioavailability (OB) and drug-likeness (DL) were used as the main indicators to identify pivotal compounds. In our study, the active compounds of Jingulian were screened according to the criterion of OB ≥30% and DL ≥0.18.
Identification of active targets
The ingredients of active compounds were collected and imported into the Swiss Target Prediction Database (swisstargetprediction.ch) (11), Batman (12), and DrugBank (http://go.drugbank.com) (13). The names and ID of the target genes were uniformly obtained using UniProtKB (http://www.uniprot.org) (14).
Identification of RA/COVID-19-associated genes
To identify the RA/COVID-19-associated genes, genes related to RA and COVID-19 were retrieved from Genecards (https://www.genecards.org/) (15), the Online Mendelian Inheritance in Man (OMIM) database (https://www.omim.org/) (16), the Therapeutic Target Database (TTD, db.idrblab.net/ttd) (17,18) and the NCBI gene module. Then, overlapping targets of RA and COVID-19 were obtained. Finally, the intersection of retrieved targets of active compounds and RA/COVID-19-related genes were then collected as the targets of Jingulian in the treatment of patients with RA/COVID-19.
Molecular docking
The three-dimensional (3D) structures of active compounds were acquired from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) (19). 3D structures of RA/COVID-19-related genes were downloaded from the Protein Data Bank (PDB) database (https://www.rcsb.org/). Autodock Vina (version 1.1.2) was used to dock small molecules. The best affinity was selected as the final docking conformation and visualized in Pymol 2.3.
Statistical analysis
The intersection genes of Jingulian and RA/COVID-19 were retrieved for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis in the DAVID database (https://david.ncifcrf.gov/) (18) with the criterion of P value >0.05. GO enrichment analysis included biological processes, molecular functions, and cellular components, and KEGG pathway analysis was performed using the ‘ClusterProfiler’, ‘GOplot’, and ‘org.Hs.eg.Db’ packages. Then, all intersected targets of active compounds and RA/COVID-19-related genes were put into the STRING database to build the protein-protein interaction (PPI) network. Cytoscape software (version 3.6.1) was used for visualization of the compound-target genes-RA/COVID19 network.
Results
Identification of Jingulian targets intersecting with RA and COVID-19
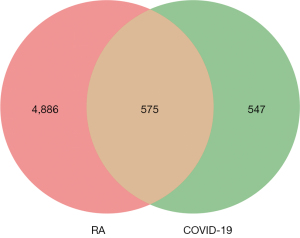
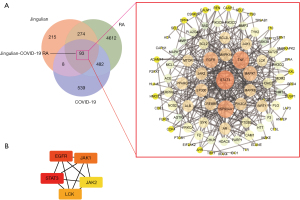
First, network pharmacology identified 5,461 genes associated with RA and 1,122 genes associated with COVID-19 from the Genecards, OMIM, TTD, and NCBI databases (Figure 2). In this study, 40 active compounds were identified through the absorption, distribution, metabolism, and excretion (ADME) screening. After biological correction and deletion of repetitive genes using the UniProt database, a total of 590 targets were identified through the TCMSP database, traditional Chinese medicine database@TAIWAN, and CNKI database using these 40 compounds. An overlap of RA/COVID-19 genes with Jingulian-associated targets identified 93 intersecting genes.
GO and KEGG enrichment
To further explore the intersecting genes, GO enrichment was conducted, which indicated that Jingulian affected a series of biological processes including protein binding, adenosine triphosphate (ATP) binding, protein kinase activity, identical protein binding, and protein serine/threonine kinase activity. After the analysis of cellular components, the targets mainly consisted of the cytoplasm, cytosol, nucleus, and plasma membrane, among others. Furthermore, molecular functions mainly included protein phosphorylation, positive regulation of transcription from RNA polymerase II promoter, signal transduction, positive regulation of cell proliferation, innate immune response, and viral process, among others (Figure 3).
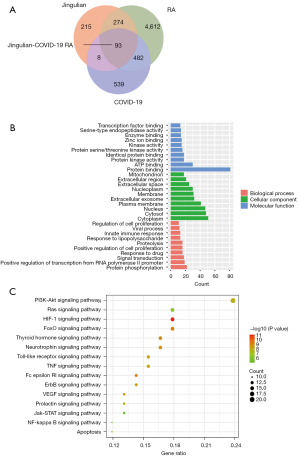
Additionally, KEGG enrichment terms related to the intersecting genes included the PI3K-Akt signaling pathway, Ras signaling pathway, HIF-1 signaling pathway, and FoxO signaling pathway, among others (Figure 3).
Identification of the main targets of Jingulian against COVID-19/RA
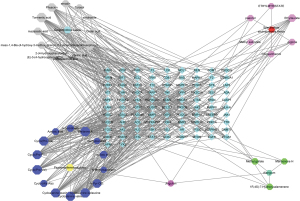
Then, STRING analysis was performed to obtain the PPI network mediated by 93 overlapped genes of Jingulian in the treatment of patients with COVID-19/RA. These intersection genes were input into Cytoscape software to determine 5 core gene targets related to Jingulian against COVID-19/RA, including signal transducer and activator of transcription 3 (STAT3), Janus kinase 1 (JAK1), epidermal growth factor receptor (EGFR), Janus kinase 2 (JAK2), and lymphocyte specific protein tyrosine kinase (LCK) (Figure 4). Finally, a Jingulian-compound-target network was constructed (Figure 5).
Results of molecular docking
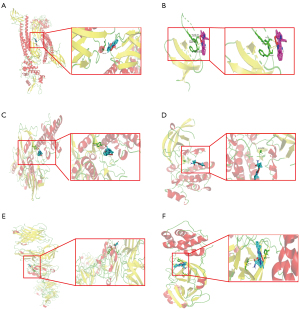
In order to explore the possible binding of ingredients of Jingulian to the targets associated with RA/COVID-19, the 5 core proteins in the PPI network were selected to perform molecular docking with the top 5 active ingredients in the drug component-target-pathway network. Among the docking results, the main binding complexes are displayed in Figure 6, including STAT3-tormentic acid docking (−11.4 kcal/mol), JAK1-tormentic acid docking (−12.6 kcal/mol), EGFR-tormentic acid docking (−12.6 kcal/mol), JAK2-tormentic acid docking (−10.3 kcal/mol), and LCK-tormentic acid docking (−8.4 kcal/mol). The 3D structure of STAT3 was obtained from the PDB database (https://www.rcsb.org/structure/6TLC), and the active cavity box parameter setting center_x, y, z was 0.231, 37.283, and 33.516, respectively. Size_x, y, z was 126, 70, and 126, respectively, whereas the RMSD of the original ligand was 2.3Å. This suggested that tormentic acid had good binding activity with the STAT3 protein. Furthermore, we obtained the 3D structure of EGFR from the PDB database (https://www.rcsb.org/structure/1YY9). The active cavity box parameter setting center_x, y, z was 35.548, 39.543, and 50.341, respectively. Size_x, y, z was 82, 72, and 126, respectively, whereas the RMSD of the original ligand was 2.7Å. The analysis suggested that hydrogen bonding was formed on 2 amino acid residues TRP-386 and SER-418 (Figure 6C), indicating the high-affinity association between tormentic acid and EGFR. In addition, we collected the crystal structure of COVID-19 main protease (https://www.rcsb.org/structure/5R84). The active cavity box parameter setting center_x, y, z was 11.891, 0.634, and 4.517, respectively. Size_x, y, z was 40, 70, and 72, respectively, indicating that tormentic acid had good binding activity with the COVID-19 main protease (Figure 6).
Discussion
The prevalence of COVID-19 has had a tremendous impact, leading to a global health emergency with high morbidity and mortality. The treatment of COVID-19 patients includes general supportive care, symptomatic treatment, and respiratory support, among others (20). However, until now, limited specific therapies against COVID-19 have been developed to overcome this disease. A study has reported that COVID-19 can lead to severe inflammation and autoimmune dysfunction (21). Meanwhile, patients with active RA face a higher risk of severe COVID-19 outcomes. Moreover, the incidence of COVID-19 in RA patients may decrease the survival rate.
In the previous reference analysis, glucocorticoid treatment was associated with a higher risk of severe COVID-19 outcomes in RA patients (22). At present, there is no specific drug for RA/COVID-19. Thus, an alternative treatment was explored in patients with COVID-19 and RA. Jingulian, a TCM mainly made from Alangium, Sargentodoxa Caulis, Speranskiae seulmpaticntis Herba, Psammosilene tunicoides, and Guangxi schefflera twig and leaf, has been used to treat RA with remarkable outcomes and few side effects, and can disperse swelling and has analgesic and anti-inflammatory effects. However, illuminating the mechanisms of TCM in treatment of RA and COVID-19 using traditional methods is challenging. Network pharmacology is a approach to understand the interaction network based on target molecules, biological function, and active compounds, which meets the natural feature of Jingulian formula, and enable elucidate the action mechanism of Jingulian at molecular level with systematic view point, expecting to be of great significance in the treatment of RA and COVID-19.
In our study, we screened out 40 active compounds of Jingulian, including protocatechuic acid (degree =103), vanillic acid (degree =26), and chrysophanol (degree =13), among others. Then, using a network pharmacology approach, 93 intersection genes were identified with Jingulian treatment against COVID-19 and RA. KEGG enrichment analysis showed that the key targets were mainly concentrated on the PI3K-Akt signaling pathway, the HIF-1 signaling pathway, the Ras signaling pathway, and the FoxO signaling pathway, among others. The results of the PPI network indicated that STAT3, JAK1, EGFR, JAK2, and LCK were the main proteins related to the ingredients of Jingulian against COVID-19/RA. PIK3CA encodes class I phosphatidylinositol-3-kinase (PI3K). A previous study has reported that the PI3K signaling pathway is related to inflammatory responses and participates in RA-related chondrocyte proliferation, autophagy, and apoptosis (23). Protocatechuic acid, a pivotal component of Jingulian, can reduce inflammation and inhibit proliferation and migration through the PI3K/AKT/mTOR pathway, so as to protect the synovial tissue of the footpad in the rheumatoid joint of rats (24). STAT3 is activated in response to the phosphorylation of various cytokines and growth factors, such as IFN-γ, IL-5, IL-6, and LIF. A previous study reported that the novel synthetic peptide AESIS-1 could be an effective therapeutic for treating RA via the downregulation of STAT3 signaling (25). Therefore, we speculated that Jingulian may directly activate the PI3K pathway and inhibits the molecular function of STAT3 to reduce the inflammatory response.
In our data, GO enrichment analysis showed that the major molecular functions were innate immune responses and viral processes. Protocatechuic acid is a phenolic compound that induces a better antiviral effect by immune enhancement. Additionally, paeonol was shown to protect against cytokine release of FLS by upregulating the FOXO signaling pathway (26). These findings indicate that the anti-RA/COVID-19 action of Jingulian may be modulated by FoxO signaling pathway. Using a molecular docking approach, we identified good binding activity between tormentic acid and STAT3, which may indicate that the compound can effectively bind to specific proteins in COVID-19. However, further clinical and experimental research is still needed.
Conclusions
Taken together, the network pharmacology analysis and molecular docking results highlight anti-inflammation and immune regulation as the main targets of Jingulian treatment in patients with COVID-19/RA. Furthermore, based on the significant pathways and targets, our studies indicated that Jingulian may be used clinically to treat patients with RA/COVID-19.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (Nos. 81904319, 82001711), the Science and Technology Fund of Guizhou Provincial Health Department (Nos. qiankehejichu-ZK[2022]zhongdian043, qiankehezhicheng[2019]Y280), the Fund of Guiyang Science and Technology Department (No. zhukehetong[2021]43-6), the Health and Family Planning Commission of Guizhou Province (Nos. gzwjkj2021-106, gzwjkj2020-1-083), the Fund of the Affiliated Hospital of Guizhou Medical University (No. gyfybsky-2021-33), the Natural Science Foundation of Jiangsu Province (No. BK20201137), and the Health and Family Planning Commission of Wuxi (No. Q202007).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1665/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1665/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Khan M, Adil SF, Alkhathlan HZ, et al. COVID-19: A Global Challenge with Old History, Epidemiology and Progress So Far. Molecules 2020;26:39. [Crossref] [PubMed]
- McCreary EK, Angus DC. Efficacy of Remdesivir in COVID-19. JAMA 2020;324:1041-2. [Crossref] [PubMed]
- Liu W, Fan Y, Tian C, et al. Deciphering the Molecular Targets and Mechanisms of HGWD in the Treatment of Rheumatoid Arthritis via Network Pharmacology and Molecular Docking. Evid Based Complement Alternat Med 2020;2020:7151634. [Crossref] [PubMed]
- Sparks JA. Rheumatoid Arthritis. Ann Intern Med 2019;170:ITC1-ITC16. [Crossref] [PubMed]
- Favalli EG, Ingegnoli F, De Lucia O, et al. COVID-19 infection and rheumatoid arthritis: Faraway, so close! Autoimmun Rev 2020;19:102523. [Crossref] [PubMed]
- Schett G, Manger B, Simon D, et al. COVID-19 revisiting inflammatory pathways of arthritis. Nat Rev Rheumatol 2020;16:465-70. [Crossref] [PubMed]
- Luo TT, Lu Y, Yan SK, et al. Network Pharmacology in Research of Chinese Medicine Formula: Methodology, Application and Prospective. Chin J Integr Med 2020;26:72-80. [Crossref] [PubMed]
- Pinzi L, Rastelli G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int J Mol Sci 2019;20:4331. [Crossref] [PubMed]
- Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:13. [Crossref] [PubMed]
- Chen CY. TCM Database@Taiwan: the world's largest traditional Chinese medicine database for drug screening in silico. PLoS One 2011;6:e15939. [Crossref] [PubMed]
- Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res 2019;47:W357-64. [Crossref] [PubMed]
- Liu Z, Guo F, Wang Y, et al. BATMAN-TCM: a Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine. Sci Rep 2016;6:21146. [Crossref] [PubMed]
- Wishart DS, Knox C, Guo AC, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res 2008;36:D901-6. [Crossref] [PubMed]
- The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res 2017;45:D158-69. [Crossref] [PubMed]
- Safran M, Dalah I, Alexander J, et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010;2010:baq020. [Crossref] [PubMed]
- Amberger JS, Bocchini CA, Schiettecatte F, et al. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 2015;43:D789-98. [Crossref] [PubMed]
- Wang Y, Zhang S, Li F, et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res 2020;48:D1031-41. [PubMed]
- Zhu F, Han B, Kumar P, et al. Update of TTD: Therapeutic Target Database. Nucleic Acids Res 2010;38:D787-91. [Crossref] [PubMed]
- Kim S, Thiessen PA, Bolton EE, et al. PubChem Substance and Compound databases. Nucleic Acids Res 2016;44:D1202-13. [Crossref] [PubMed]
- Seyed Hosseini E, Riahi Kashani N, Nikzad H, et al. The novel coronavirus Disease-2019 (COVID-19): Mechanism of action, detection and recent therapeutic strategies. Virology 2020;551:1-9. [Crossref] [PubMed]
- Mohammadi S, Moosaie F, Aarabi MH. Understanding the Immunologic Characteristics of Neurologic Manifestations of SARS-CoV-2 and Potential Immunological Mechanisms. Mol Neurobiol 2020;57:5263-75. [Crossref] [PubMed]
- Raiker R, DeYoung C, Pakhchanian H, et al. Outcomes of COVID-19 in patients with rheumatoid arthritis: A multicenter research network study in the United States. Semin Arthritis Rheum 2021;51:1057-66. [Crossref] [PubMed]
- Feng FB, Qiu HY. Effects of Artesunate on chondrocyte proliferation, apoptosis and autophagy through the PI3K/AKT/mTOR signaling pathway in rat models with rheumatoid arthritis. Biomed Pharmacother 2018;102:1209-20. [Crossref] [PubMed]
- Wu H, Wang J, Zhao Q, et al. Protocatechuic acid inhibits proliferation, migration and inflammatory response in rheumatoid arthritis fibroblast-like synoviocytes. Artif Cells Nanomed Biotechnol 2020;48:969-76. [Crossref] [PubMed]
- Kim KE, Jeon S, Song J, et al. The Novel Synthetic Peptide AESIS-1 Exerts a Preventive Effect on Collagen-Induced Arthritis Mouse Model via STAT3 Suppression. Int J Mol Sci 2020;21:378. [Crossref] [PubMed]
- Liu N, Feng X, Wang W, et al. Paeonol protects against TNF-α-induced proliferation and cytokine release of rheumatoid arthritis fibroblast-like synoviocytes by upregulating FOXO3 through inhibition of miR-155 expression. Inflamm Res 2017;66:603-10. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


