
Identification of novel drug targets for the risk and prognosis of COVID-19
Introduction
Known as coronavirus disease 2019 (COVID-19), the pneumonia caused by SARS-CoV-2 infection has spread to 220 countries with 153,187,889 confirmed cases including 3,209,109 deaths reported to World Health Organization (WHO) as of 4:20 pm CEST, 4 May 2021, leading to widespread social and economic disruption (1). With a wide spectrum of clinical manifestations, high heterogeneity in both susceptibility and severity of SARS-CoV2 infection was shown. Common symptoms of COVID-19 include fever or chills, cough, headache et cetera, while severe cases can have shortness of breath or difficulty breathing, persistent pain or pressure in the chest, confusion, inability to wake or stay awake and even acute respiratory distress syndrome (ARDS).
To date, a number of vaccines against SARS-CoV-2 have been licensed and used. However, it still takes time to achieve the aim of 70 percent of the world population being vaccinated, and vaccine to prevent multi-generational transmission of the virus has not yet been developed. It remains unknown how long the immunity can be formed after infection with the virus or vaccination. Thus, to develop new and better treatments against the disease is still necessary. Mendelian randomization (MR) is a strategy for assessing the causal effect of modifiable exposures on disease using human genetic variation known to influence the exposures. With the introduction of instrumental variables (IV), which are innately determined genetic variants, MR offers a way to avoid the influence of confounding factors. Within detection for circulating proteins that correlate with susceptibility and severity of SARS-CoV2 infection, we may be able to find new perspectives on the pathological mechanism of the disease, and possible drug targets for treatment. This becomes feasible since recent technological advances in high-throughput protein quantification have enabled genome-wide association studies (GWAS) of genetic determinants of blood proteins, and COVID-19 GWAS meta-analyses are being performed worldwide.
Here in this study, we used MR approach to assess the relationship between circulating proteins derived from six biomarker GWAS analyses and COVID-19 susceptibility and severity. We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6612/rc).
Methods
Characterizing genetic instruments for proteins
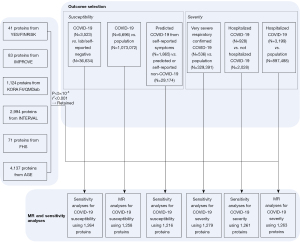
Briefly, we combined six different biomarker GWAS analyses, of which genome-wide summary statistics were publicly available [Cardiovascular Risk in Young Finns Study/FINRISK (YFS/FINRISK) (2); IMPROVE (3); INTERVAL (4); FHS (5); AGES (6); KORA F4/QMDiab (7)]. Genetic determinants of circulating biomarker levels were derived. Biomarker testing was conducted in blood samples in the six study samples consisted of Caucasians. Various high-multiplex protein assays were used to conduct biomarker testing in the six study samples. Specifically, YFS/FINRISK analyzed 41 cytokines via bead-based immunoassays. IMPROVE analyzed 83 proteins via modified antibodies conjugated to ligonucleotides. FHS analyzed 71 proteins via modified enzyme-linked immunosorbent assay sandwich method. KORA F4/QMDiab, INTERVAL and AGES analyzed 1,124 proteins, 2,994 proteins and 4,137 proteins respectively via SOMAmers. For further details, please refer to Table S1. Among all the biomarkers, uncorrelated (r2<0.001) single-nucleotide polymorphisms associated with the corresponding exposure trait at genome-wide significance (P<5×10−8) were retained as instrumental variables. Information of all the included identified biomarkers are available in Table S1.
Characterizing COVID-19 susceptibility and severity
The COVID-19 Host Genetics Initiative where we extracted the data from is a bottom-up collaborative aims to provide a platform for sharing resources, organizing analytical activities and sharing results of such studies to identify genetic determinants of COVID-19 susceptibility and severity. Summary statistics from the third round of GWAS meta-analysis, shared publicly on July 2, 2020, which were available via the platform, were used to test the genetic instruments aforementioned against COVID-19 outcomes.
For our two primary analyses, we selected two samples with the largest number of cases from the above platform. For susceptibility analysis, we chose COVID-19 positive patients diagnosed by RNA PCR, serologic testing, or clinician diagnosis by chart review or ICD-coding (N=6,696) vs. population controls (N=1,073,072). And for severity analysis, we chose Hospitalized COVID-19 positive patients diagnosed by RNA PCR, serologic testing, or clinician diagnosis by chart review or ICD-coding (N=3,199) vs. population controls (N=897,488). The population controls were defined as any person who was not a case, which means who were tested negative, were never tested, or had an unknown testing status.
Four remained available outcomes from the platform were used to determine whether statistically significant results from the primary analyses were consistent across different definitions for COVID-19 susceptibility, severity, and control groups. For susceptibility: (I) COVID-19 positive by RNA PCR, serologic testing, or clinician diagnosis by chart review or ICD-coding (N=3,523) vs. lab/self-reported negative (N=36,634); (II) predicted COVID-19 from self-reported symptoms (N=1,865) vs. predicted or self-reported non-COVID-19 (N=29,174). For severity: (III) critical cases of COVID-19 defined by death, intubation, continuous positive airway pressure (CPAP), bilevel positive airway pressure (BiPAP), continued external negative pressure (CNP), or very high flow positive end expiratory pressure oxygen in patients with COVID-19 by RNA PCR or serologic testing (N=536) vs. population control (N=329,391); (IV) hospitalized COVID-19 positive (N=928) vs. non-hospitalized COVID-19 positive (N=2,028). The information of the above COVID-19 GWAS is available in Table S2.
Statistics analyses
The Wald ratio method was chosen for estimating the causal association between each exposure with each outcome since most of the exposures contained only one SNP. Suppose we have an IV (we can think of IV as a single nucleotide polymorphism) with value of 1 or 0, the entire population can be divided into two genetic groups based on this. Two of the three subgroups can be combined according to a dominant or recessive model, or if there are only a few individuals in a genetic subgroup (minor homozygotes), it can also be combined. According to the hypothesis, if both exposure distributions and outcome distributions of the two genetic subgroups differs from each other, it supports that the exposure has a causal relationship with the outcome. The ratio is the coefficient of the genetic variant in the regression of the outcome divided by the coefficient of the genetic variant in the regression of the exposure (8). Other methods including inverse variance weighted (IVW), MR-egger and median weighted MR were also employed.
A Bonferroni-corrected P value threshold accounting for both the number of biomarkers and outcomes analyzed was implemented [since two outcomes were used in our primary analyses, we set P=1.98×10−5=0.05/(1,263×2)]. We defined significant results as those with P<1.98×10−5 (after Bonferroni correction), and suggestive associations as those with 1.98×10−5<P<0.05. We then performed standard sensitivity analyses including Wald ratio method and the others to assess the validity of the MR findings. Each exposure was tested with each outcome like the above operation. All the MR testing were performed using the “MRBase for TwoSample MR” package (version 0.4.09). The detailed process is available in Figure 1.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The current analyses are based on publicly available summary data and therefore do not require ethical approval. Original studies have been approved by ethic committees and written informed consent was obtained from study participants or caregivers.
Results
Selection of genetic instruments for exposures
For our primary analyses, after excluding variants that r2>0.001 and those with weak P values (P>5×10−8), 1,258 proteins were tested for the correlation with COVID-19 susceptibility, and 1,263 proteins were estimated for whether to be causally associated with COVID-19 severity. Contributing studies included in these exposure GWAS meta-analyses were predominantly of Caucasians.
Causal effect of each exposure on COVID-19 susceptibility and severity
Significant results were defined as those with P<1.98×10−5 (after Bonferroni correction), whereas those with 1.98×10−5<P<0.05 were defined as suggestive associations. MR results were presented in Figures 2,3. Traits were shown in the figure if Wald ratio or MR-IVW or MR-Weighted-Median showed nominally significant (P<0.05) results. Detail information is available in Table S3.
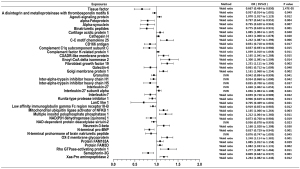
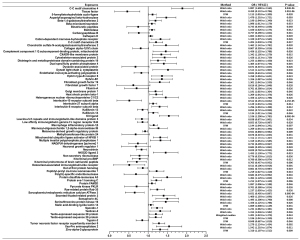
Of all the exposures, we observed tissue factor (TF) to be causally associated with COVID-19 susceptibility, while both TF and C-C motif chemokine 4 were found to have causal relationship with COVID-19 severity. Specifically, for susceptibility, we found that TF was negatively related to diagnosis of COVID-19 (Wald ratio, OR =0.667, 95% CI: 0.484–0.850, P=1.47×10−5). For severity, we found that TF was significantly associated with lower risk of hospitalization (Wald ratio, OR =0.459, 95% CI: 0.132–0.786, P=3.01×10−6). Whereas C-C motif chemokine 4 showed association with higher risk of COVID-19 hospitalization (Wald ratio, OR =1.887, 95% CI: 1.608–2.165, P=8.04×10−6).
The evidence indicated that TF might be a protective factor of COVID-19, while C-C motif chemokine 4 might be a risk factor. Other 38 proteins were exhibited suggestive association with COVID-19 susceptibility whereas 70 proteins were exhibited suggestive association with COVID-19 severity. The complete information is available in Table S3.
Sensitivity analyses
To demonstrate whether different definition of COVID-19 susceptibility and severity and control groups would influence on the results, we then repeated the analyses using the four other outcomes obtained from the COVID-19 Host Genetics Initiative.
As aforementioned, for susceptibility, 1,264 proteins were tested with the outcome of COVID-19 positive cases diagnosed by RNA PCR, serologic testing, or clinician diagnosis by chart review or ICD-coding vs. lab/self-reported negative cases (Bonferroni correction: P=3.96×10−5=0.05/1,264, those with P<3.96×10−5 were defined as significant results), and 1,216 proteins were tested with the outcome of predicted COVID-19 cases from self-reported symptoms vs. predicted or self-reported non-COVID-19 cases (Bonferroni correction: P=4.11×10−5=0.05/1,216, those with P<4.11×10−5 were defined as significant results). For severity, 1,279 proteins were tested with the outcome of critical cases of COVID-19 vs. population control (Bonferroni correction: P=3.91×10−5=0.05/1,279, those with P<3.91×10−5 were defined as significant results), and 1,261 proteins were tested with the outcome of hospitalized COVID-19 positive cases vs. non-hospitalized COVID-19 positive cases (Bonferroni correction: P=3.97×10−5=0.05/1,261, those with P<3.97×10−5 were defined as significant results). MR results of sensitivity analyses are presented in https://cdn.amegroups.cn/static/public/atm-21-6612-1.docx.
Of all the exposures, only 2'-5'-oligoadenylate synthase 1 was found to be negatively associated with increased risk of confirmed COVID-19 with critical respiratory illness (IVW, OR =0.511, 95% CI: 0.266–0.757, P=8.51×10−8). Other 107 proteins were exhibited suggestive association with COVID-19 susceptibility whereas 124 proteins were exhibited suggestive association with COVID-19 severity. The complete information is available in https://cdn.amegroups.cn/static/public/atm-21-6612-1.docx.
Summary findings
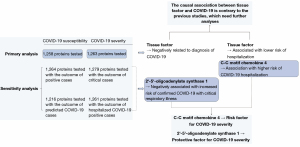
In summary, association of three circulating proteins with COVID-19 susceptibility and severity were demonstrated via our study. The most consistent finding was tentative evidence which revealed C-C motif chemokine 4 as a risk factor for COVID-19 severity, and 2'-5'-oligoadenylate synthase 1 as a protective factor for COVID-19 severity. TF was found to be a possible protective factor for both COVID-19 susceptibility and severity, which was contrary to the previous studies, needing further analyses. A brief summary of the findings is present in Figure 4.
Discussion
In this study, MR was applied to estimate the causal association between blood proteome and COVID-19 susceptibility and severity. In our primary analyses, 1,258 circulating proteins were tested for the correlation with COVID-19 susceptibility, and 1,263 proteins were tested for the correlation with COVID-19 severity. TF was identified as a possible protective factor for both COVID-19 susceptibility and severity, whereas C-C motif chemokine 4 was identified as a risk factor for COVID-19 severity. MR was further applied to perform sensitivity analyses, and 2'-5'-oligoadenylate synthase 1 was found to be negatively correlated with COVID-19 severity. The different proteins we found causally correlated with COVID-19 susceptibility and severity in our primary analyses and sensitivity analyses indicate that different definition of COVID-19 susceptibility and severity might have influence on the result of the analyses.
The evidences revealed the causal relationship between C-C motif chemokine 4 and 2'-5'-oligoadenylate synthase 1 and COVID-19 severity are the most important findings of this study. As no previous study has shown that C-C motif chemokine 4 is correlated to COVID-19 severity, our study first indicates that increase of blood C-C motif chemokine 4 leads to a higher risk of COVID-19 hospitalization, and the possible mechanism might be related to the role it plays in the overexpression of inflammatory factors and inflammatory injury in the lungs. C-C motif chemokine 4 expresses in granulocyte and 180 other tissues. The function of the chemotactic cytokine family it belongs to is to induce direct chemotaxis in nearby responsive cells, recruiting cells of the immune system to a site of infection during immune response (9). The chemokine signal is transduced by G-protein coupled receptors expressed on the immune cells. Receptor activation leads to the dissociation of the α and β-γ-subunits of G protein, activating diverse downstream pathways such as Jak-STAT signaling pathway, MAPK signaling pathway PLC/PKC signaling pathway and etcetera, resulting in cellular growth and differentiation, cellular polarization, apoptosis and degranulation, NO induction and ROS production, and actin reorganization (refer to chemokine signaling pathway map in KEGG: Kyoto Encyclopedia of Genes and Genomes). We suppose that C-C motif chemokine 4 participates in the aggravation of patients’ condition with COVID-19 through these pathways, since COVID-19 is characterized by an overexuberant inflammatory response, and excessive level of oxidative stress has been found in critically ill patients with COVID-19 (10). In fact, anti-inflammatory treatments have been applied in clinical trials. Previous study has found that Baricitinib, as a selective JAK inhibitor, has both antiviral and anti-inflammatory properties via a particularly high affinity for AAK1 and a vital regulator of clathrin-mediated endocytosis, while other JAK inhibitors do not have the predicted inhibition of clathrin-mediated endocytosis at a dose that patient can tolerate (11). Our study confirms the causal association between the C-C motif chemokine 4 and the severity of the disease, which indicates other possible therapeutic targets among the downstream pathways aforementioned besides Jak-STAT signaling pathway.
The critical function of oligoadenylate synthetases (OAS)/RNase L system in antiviral defense is well known, and our study demonstrates that it also plays its part during the process of COVID-19. 2'-5'-oligoadenylate synthase 1 is an interferon-induced, dsRNA-activated antiviral enzyme playing an important role in cellular innate antiviral response (12,13). The previous study has shown that 2'-5'-oligoadenylate synthase 1 has the function of anti-respiratory-syncytial-virus infection via interferon-gamma inhibition (14), and displays antiviral effect against vesicular stomatitis virus (VSV), herpes simplex virus type 2 (HSV2), and encephalomyocarditis virus (EMCV) via the classical RNase L-dependent pathway or an alternative antiviral pathway independent of RNase L. Our study indicates that high level of 2'-5'-oligoadenylate synthase 1 leads to a decreased risk of COVID-19 positive with critical respiratory illness. Refer to the previous study, we can speculate that the possible mechanism of how 2'-5'-oligoadenylate synthase works in the anti-SARS-CoV-2 response is that activated OAS catalyzes the oligomerization of ATP into 2',5'-linked oligoadenylate (2-5A) which can bind to and activate the latent RNase L (15). Activated RNase L then restricts viral propagation through both direct and indirect mechanisms including viral genome degradation, viral mRNA degradation, cellular mRNA and rRNA degradation and amplification of IFN signaling (15). Thus, this finding indicates potential implication of OAS1 activity as therapeutic target in critically ill COVID-19 patients.
The previous study on the inhibition of OAS/RNase L system by other viruses, on the other hand, remind us the possibility and potential mechanism how SARS-CoV-2 may counteract the antiviral activity of OAS/RNase L. Several methods are known used by other viruses at either upstream or downstream of the pathway, including dsRNA sequestration by a certain viral protein, expression of viral mRNA decapping enzymes, 2-5A degradation by a viral phosphodiesterase, production of inactive or inhibitory 2-5A, increased RLI/ABCE expression, inhibition of RNase L activation through direct binding to the enzyme, competitive inhibition of ribonuclease activity, and escape from RNase L cleavage through genome adaptation (14). Those possible escape mechanisms should be taken into account during future development of anti-SARS-CoV-2 treatments.
The other tentative finding of our study is that TF might be a protective factor of both COVID-19 susceptibility and severity, which is contrary to previous studies. TF is a transmembrane glycoprotein found express high level in bronchial mucosa and alveolar epithelial cells in the lungs. Following inflammatory injury in the lung, combined with FVII(a), TF is known to be correlated with the expression of several immunoregulatory genes in the lung and fibrin formation, coupled with increased cytokine production and cell migration and activation, leading to acute lung injury, for example, acute respiratory distress syndrome (ARDS) (16).
In contrary to our study, evidences in previous studies support that TF is positively related to the severity of COVID-19. Previous studies detected increased level of TF activity in COVID-19, which was correlated with the inflammatory injury and fibrin formation in the lungs of the COVID-19 patients, associated with COVID-19 severity and mortality (17,18). The previous cohort study demonstrated increased platelet activation and platelet-monocyte aggregate formation in severe COVID-19 patients, inducing TF expression in monocytes. Increased platelet activation and monocyte TF expression were associated with higher fibrinogen and D-dimers level in severe cases and could be inhibited by platelet P-selectin neutralization or integrin αIIb/β3 blocking with the aggregation inhibitor abciximab (10). Another study demonstrated higher TF expression in neutrophils in severe cases, which could be disrupted by complement C3 inhibition with compstatin Cp40, and showed that thrombotic activity of HAECs was induced by TF-bearing NETs (9).
Our study, however, shows a total different result, demonstrates the causal association between blood TF level and the decrease of COVID-19 susceptibility and severity, indicating that TF might be a protective factor of COVID-19. The possible explanation is that we might have not ruled out a clear effect of TF on COVID-19 owing to the low variance explained by only 1 genetic instrument. Whether TF has such causal association with COVID-19 susceptibility and severity need more genetic instruments to carry out further analyses.
Our study findings have several implications. First, anti-inflammatory treatment is supported since the pro-inflammatory protein (C-C motif chemokine 4) is found causally associated with the severity of COVID-19. Second, additional drug targets may be uncovered as biomarker testing becomes more comprehensive. Since OAS1 is found to be a protective factor of COVID-19 severity, it can be used as a target for further development of anti-virus drugs.
The interpretation and generalizability of study findings are limited by several factors. First, most study participants were Europeans due to limitations in data availability. Thus, further study based on non-European biomarker GWAS is necessary. Second, in our primary analyses, those who were broadly defined as not being a case were chosen to be controls. However, the control group could have been contaminated with people who had contracted COVID-19, particularly those with only mild or no viral symptoms without universal testing, which may influence the estimates in some degree. Third, the low variance explained by only 1 genetic instrument might make it hard for us to rule out a clear association between some of the blood proteins and COVID-19 susceptibility and severity. Forth, MR itself has certain limitations. The IVs are hypothesized to satisfy 3 assumptions: the IV is associated with the exposure; the IV is not associated with confounders; and the IV influences the outcome only through the exposure (19). However, due to the existence of complex biological effects, pleiotropy of the variants is usually inevitable. By using multivariable mendelian randomization (MVMR), the causal effects of each of the confounding factors can be estimated (20). However, we don’t have access to clinical data needed, so we are unable to discuss their influence on the study results.
Conclusions
Systematic MR analysis of the circulating proteome revealed C-C motif chemokine 4 as a risk factor for COVID-19 severity, and 2'-5'-oligoadenylate synthase 1 as a protective factor for COVID-19 severity. The causal association between TF and COVID-19 is contrary to the previous studies, which need further analyses. Further research is warranted to assess the viability of C-C motif chemokine 4 and 2'-5'-oligoadenylate synthase 1 as well as their downstream pathways as drug targets for anti-inflammatory and anti-virus treatment in severe cases.
Acknowledgments
We would like to acknowledge the important contributions of the following datasets which made this study possible. We acknowledge the contributions of YFS/FINRISK, IMPROVE, INTERVAL, FHS, AGES, and KORA F4/QMDiab for providing public available summary statistics of blood proteins. We thank sincerely the studies in COVID-19 Host Genetics Initiative listed in https://www.covid19hg.org/acknowledgements/.
Funding: This study was supported by grants from the Zhangjiang Lab, Tianqiao and Chrissy Chen Institute, and the State Key Laboratory of Neurobiology and Frontiers Center for Brain Science of Ministry of Education, Fudan University.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6612/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6612/coif). JTY serves as an unpaid Associate Editor-in-Chief of Annals of Translational Medicine from June 2019 to May 2024. The other authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int
- Ahola-Olli AV, Würtz P, Havulinna AS, et al. Genome-wide Association Study Identifies 27 Loci Influencing Concentrations of Circulating Cytokines and Growth Factors. Am J Hum Genet 2017;100:40-50. [Crossref] [PubMed]
- Folkersen L, Fauman E, Sabater-Lleal M, et al. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet 2017;13:e1006706. [Crossref] [PubMed]
- Sun BB, Maranville JC, Peters JE, et al. Genomic atlas of the human plasma proteome. Nature 2018;558:73-9. [Crossref] [PubMed]
- Yao C, Chen G, Song C, et al. Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat Commun 2018;9:3268. [Crossref] [PubMed]
- Emilsson V, Ilkov M, Lamb JR, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 2018;361:769-73. [Crossref] [PubMed]
- Suhre K, Arnold M, Bhagwat AM, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun 2017;8:14357. [Crossref] [PubMed]
- Rasooly D, Patel CJ. Conducting a Reproducible Mendelian Randomization Analysis Using the R Analytic Statistical Environment. Curr Protoc Hum Genet 2019;101:e82. [Crossref] [PubMed]
- Bagri A, Gurney T, He X, et al. The chemokine SDF1 regulates migration of dentate granule cells. Development 2002;129:4249-60. [Crossref] [PubMed]
- Laforge M, Elbim C, Frère C, et al. Tissue damage from neutrophil-induced oxidative stress in COVID-19. Nat Rev Immunol 2020;20:515-6. [Crossref] [PubMed]
- Stebbing J, Phelan A, Griffin I, et al. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect Dis 2020;20:400-2. [Crossref] [PubMed]
- Eskildsen S, Justesen J, Schierup MH, et al. Characterization of the 2'-5'-oligoadenylate synthetase ubiquitin-like family. Nucleic Acids Res 2003;31:3166-73. [Crossref] [PubMed]
- Lin RJ, Yu HP, Chang BL, et al. Distinct antiviral roles for human 2',5'-oligoadenylate synthetase family members against dengue virus infection. J Immunol 2009;183:8035-43. [Crossref] [PubMed]
- Behera AK, Kumar M, Lockey RF, et al. 2'-5' Oligoadenylate synthetase plays a critical role in interferon-gamma inhibition of respiratory syncytial virus infection of human epithelial cells. J Biol Chem 2002;277:25601-8. [Crossref] [PubMed]
- Drappier M, Michiels T. Inhibition of the OAS/RNase L pathway by viruses. Curr Opin Virol 2015;15:19-26. [Crossref] [PubMed]
- Laurent GJ, Shapiro SD. Encyclopedia of Respiratory Medicine. American: Academic Press, 2006:494-8.
- Skendros P, Mitsios A, Chrysanthopoulou A, et al. Complement and tissue factor-enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J Clin Invest 2020;130:6151-7. [Crossref] [PubMed]
- Hottz ED, Azevedo-Quintanilha IG, Palhinha L, et al. Platelet activation and platelet-monocyte aggregate formation trigger tissue factor expression in patients with severe COVID-19. Blood 2020;136:1330-41. [Crossref] [PubMed]
- Emdin CA, Khera AV, Kathiresan S. Mendelian Randomization. JAMA 2017;318:1925-6. [Crossref] [PubMed]
- Burgess S, Freitag DF, Khan H, et al. Using multivariable Mendelian randomization to disentangle the causal effects of lipid fractions. PLoS One 2014;9:e108891. [Crossref] [PubMed]


