Genetics of the acute coronary syndrome
Introduction
Acute coronary artery disease (CAD) is the most common cause of death in industrialized countries, accounting for up to 40% of all deaths. Moreover, it is also rapidly increasing in prevalence in developing countries due to the epidemiological and demographic changes attributable to the improvement of the socio-economic and health conditions. Indeed, as a consequence of the reduction of childhood mortality, an increasing number of individuals reach an older age, thus becoming at greater risk of cardiovascular disease (1-3).
As CAD is becoming a leading public health problem around the globe, it is not surprising that a vast part of the medical research is now focusing on the identification of genetic and environmental factors contributing to the development of this multifactorial disease (4-8). However, although the recognition and treatment of established risk factors for coronary heart disease is crucial to reduce the disease burden, studies aimed to unravel the genetic basis of coronary disease are equally important as they permit the development of novel diagnostic and therapeutic methods (9,10).
The current knowledge about the genetic determinants of acute CAD will be explored in this review.
Candidate gene studies
Acute coronary syndrome is the most important example of a complex, or multifactorial, disease. Contrary to monogenetic diseases (i.e., haemophilia or cystic fibrosis), complex diseases are not caused by a single gene defect but by variable combinations of gene-gene and gene-environment interactions (11).
While single gene disorders are considered to be rare and deterministic, a complex trait such as CAD is common and probabilistic in phenotypic determination. Indeed, complex diseases do not follow a clear pattern of Mendelian inheritance (i.e., autosomal dominant, recessive or X-linked) but are characterized by the lack of a perfect co-segregation between the risk allele and phenotypic manifestation and by a high prevalence in the population (11). Thus, in a complex trait, the presence of a risk allele is neither necessary nor sufficient to cause the phenotype and, hence, establishing causality is particularly demanding (12). As a consequence, genetic studies on multifactorial diseases have been difficult to perform.
Candidate gene case-control association studies have been identified as the most used and easiest method to identify the susceptibility genes for CAD. By means of this approach, single-nucleotide polymorphisms (SNPs), occurring at a frequency of greater than 1% in the general population, are identified in the candidate gene and genotyped in a group of patients (cases) and in controls. The frequencies of SNP alleles or genotypes are then analysed, and an association between the allele/genotype and the disease is established if its occurrence in the cases is statistically different from that in the controls (13,14). Numerous case-control association studies were carried out over the past decade, and variants in many genes have been implicated in increasing or decreasing the susceptibility to coronary heart disease. However, the results of the different studies were quite conflicting as documented by a number of recent reviews and meta-analyses (13). Among the most convincingly replicated findings is that for the ε4 allele of apolipoprotein E, which may be associated with a 1.4-fold increased risk of CAD according to a meta-analysis by Song and colleagues (15). Among the hemostasis gene variants, only the gain-of-function factor V G1691A and prothrombin G20210A mutations were associated with a modestly increased risk [odds ratio (OR) of 1.2–1.3] in the frame of a huge meta-analysis published by Ye and colleagues (16).
Significant advances have been made in identifying chromosomal loci linked to or genetic variations that confer susceptibility to CAD using microarray-based or high-throughput RNA sequencing methods (17,18). The GENEQUEST was the first study that applied these newer techniques to the genetic investigation of CAD (19). The study analysed the relationship between 72 SNPs from 62 candidate genes and CAD in 362 case subjects (patients with at least one affected sibling with premature CAD) and 402 control subjects. Among all the SNPs studied, the most significant association with premature myocardial infarction was found with three variants in the thrombospondin gene (THBS1, THBS2 and THBS4), which encodes for a family of glycoproteins considered to play an act as modulators of cell-matrix interactions and angiogenesis. Several other large-scale SNP studies have described additional polymorphisms associated with CAD. Yamada and colleagues used a two-step approach to select for polymorphisms associate with myocardial infarction (20). They initially screened 112 polymorphisms from 71 candidate genes in 909 unrelated patients, and found 19 polymorphisms in men and 18 in women associated with myocardial infarction. They then confirmed the significance of these candidate SNPs through a case control study of 2,819 Japanese patients with myocardial infarction and 2,242 control subjects. With this approach, they described an increased risk of myocardial infarction associated with C1019T polymorphism in the Connexin 37 gene (OR, 1.4) in men and a 4G-668/5G polymorphism in the plasminogen activator inhibitor type 1 (PAI-1) gene (OR, 1.6) and a 5A-1171/6A polymorphism in the stromelysin-1 gene (OR, 4.7) in women. McCarthy and colleagues extended the initial results from GENEQUEST by screening 210 polymorphisms in 111 candidate genes from 418 patients with premature CAD (21). They found strong associations with the previously described variants of THBS2 (OR, 0.38) and THBS4 (OR, 2.22) and plasminogen activator inhibitor-2 (PAI2) (OR, 2.35). Several other novel genes related to diverse biochemical and cellular functions involved in the pathogenesis of CAD were identified by a more recent large and systematic candidate gene study (22).
Genome wide association studies
In addition to high-throughput genotyping, genome-wide scanning has been more recently introduced to identify potential genes for further analysis. Genome-wide association studies (GWAS) are large-scale population-based studies that utilize chip technology for the identification of association between genetic markers and particular clinical phenotypes (23,24). A genome-wide association approach has become possible with data from the International HapMap Project, which was built to create a public resource of common SNPs to capture most of the common human genome sequence variability ant thus provided a powerful resource of markers for genome-wide association studies (25). The first genome-wide association study was published by Ozaki and colleagues in Japan in 2002, analyzing more than 65,000 SNPs in nearly 14,000 genes (26). Six million genotypes were generated and SNPs identified as associated with myocardial infarction were then typed in a separate case series. A significant association was observed between myocardial infarction and lymphotoxin-alpha gene (LTA) which, interestingly, encodes a member of the tumor necrosis factor (TNF) ligand family and has been implicated in atherosclerosis in animal models (27). However, the first genetic risk variant with a robust association with CAD was identified by three independent groups on the short arm of chromosome 9 (9p21.3) (28-30). McPherson and colleagues screened 100,000 SNPs throughout the genome, and found that two SNPs (rs10757274 and rs2383206) located within 20 kilobases of each other on the chromosome 9p21 were associated with the risk of CAD in a Canadian population. These findings were then replicated in five other large Caucasian cohorts from Denmark and United States (28). Helgadottir and colleagues screened 305,953 SNPs, describing a significant association between myocardial infarction risk and SNPs on chromosome 9p21 (rs2383207 and rs10757278) in an Icelandic population, followed by replication in four other US Caucasian cohorts (29). The same susceptibility locus was also identified in the frame of a genome-wide association study of 1,926 cases and 3,000 controls from a British population by the Wellcome Trust Case Control Consortium (31), and replicated in a German population (30). The association between CAD and these SNPs was also replicated in several other independent studies, such as the Framingham Heart Study (32) and the PROCARDIS Consortium which included 4,251 cases with CAD and 4,443 controls from four European populations (33), thus identifying this locus as the most highly replicated for the risk of CAD. Interestingly, the chromosome 9p21 region contains genes coding for two cyclin-dependent kinase inhibitors, CCDKN2A and CDKN2B, which are known to play critical roles in cell proliferation, aging, senescence and apoptosis (34). Since then, several other loci have been identified by other study groups. For instance, the international consortium designed Coronary Artery Disease Genome-Wide Replication and Meta-Analysis (CARDIoGRAM) study, which involved investigators from the UK, Germany, USA, Iceland and Canada, led to the discovery of 13 new genetic risk variants for CAD, with confirmation of 10 previously identified risk variants (35). Notably, in another study involving CARDIoGRAM investigators, Reilly and colleagues identified a protective role against myocardial infarction for several SNPs tagging the O allele at the ABO locus at chromosome 9p34.2 (36). While A and B alleles encode glycosyltransferases that transfer carbohydrates to von Willebrand factor (VWF), the O allele encodes a protein without any enzyme activity which is thought to facilitate the proteolysis of VWF with resulting lower circulating VWF and factor VIII and reduced risk of coronary thrombosis (37). The National Human Genome Research Institute (NHGRI) Catalog has currently included 29 genome wide association studies, which identified more than 150 genomic loci associated with CAD in different populations (38). Collectively, these data document that the susceptibility to this common disease is largely determined by common SNPs of small effect size (OR in the range 1.1–1.3, with the exception of the LPA gene at the locus 9p21 encoding for lipoprotein (a) and a region of unknown function at 6p24) but with genome wide significance (P<5×10−8) (39). Table 1 summarizes the main chromosomal loci for CAD identified by genome wide association studies.
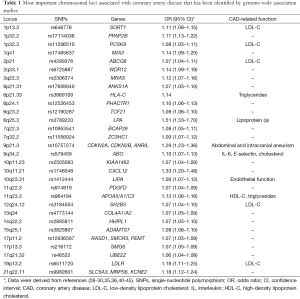
Full table
Conclusions
CAD is a complex multifactorial disease which has, however, an important genetic component. Indeed, thanks to recent biotechnological progresses, the role of inheritance in the risk of developing CAD has been defined more accurately. Genome wide association studies have definitely elucidated that such disease is the result of the interaction of common genetic variants, mostly implicated in the lipid metabolism, cell proliferation and inflammation (Table 1). While these polymorphisms have individually a small effect on the CAD risk, they are closely associated each other in a synergic manner, thus accounting for the high incidence of this clinical condition, which is currently the leading cause of morbidity and mortality in high income countries.
Acknowledgements
None.
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Breslow JL. Cardiovascular disease burden increases, NIH funding decreases. Nat Med 1997;3:600-1. [Crossref] [PubMed]
- Okrainec K, Banerjee DK, Eisenberg MJ. Coronary artery disease in the developing world. Am Heart J 2004;148:7-15. [Crossref] [PubMed]
- Thom T, Haase N, Rosamond W, et al. Heart disease and stroke statistics--2006 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 2006;113:e85-151. [Crossref] [PubMed]
- Wang Q. Advances in the genetic basis of coronary artery disease. Curr Atheroscler Rep 2005;7:235-41. [Crossref] [PubMed]
- Kullo IJ, Ding K. Mechanisms of disease: The genetic basis of coronary heart disease. Nat Clin Pract Cardiovasc Med 2007;4:558-69. [Crossref] [PubMed]
- Topol EJ, Smith J, Plow EF, et al. Genetic susceptibility to myocardial infarction and coronary artery disease. Hum Mol Genet 2006;15:R117-23. [Crossref] [PubMed]
- Franchini M, Peyvandi F, Mannucci PM. The genetic basis of coronary artery disease: from candidate genes to whole genome analysis. Trends Cardiovasc Med 2008;18:157-62. [Crossref] [PubMed]
- Lippi G, Franchini M, Targher G. Arterial thrombus formation in cardiovascular disease. Nat Rev Cardiol 2011;8:502-12. [Crossref] [PubMed]
- Lippi G, Franchini M, Cervellin G. Diagnosis and management of ischemic heart disease. Semin Thromb Hemost 2013;39:202-13. [Crossref] [PubMed]
- Delles C, McBride MW, Padmanabhan S, et al. The genetics of cardiovascular disease. Trends Endocrinol Metab 2008;19:309-16. [Crossref] [PubMed]
- Ginsburg GS, Donahue MP, Newby LK. Prospects for personalized cardiovascular medicine: the impact of genomics. J Am Coll Cardiol 2005;46:1615-27. [Crossref] [PubMed]
- Nabel EG. Cardiovascular disease. N Engl J Med 2003;349:60-72. [Crossref] [PubMed]
- Cambien F, Tiret L. Genetics of cardiovascular diseases: from single mutations to the whole genome. Circulation 2007;116:1714-24. [Crossref] [PubMed]
- Wang Q. Molecular genetics of coronary artery disease. Curr Opin Cardiol 2005;20:182-8. [Crossref] [PubMed]
- Song Y, Stampfer MJ, Liu S. Meta-analysis: apolipoprotein E genotypes and risk for coronary heart disease. Ann Intern Med 2004;141:137-47. [Crossref] [PubMed]
- Ye Z, Liu EH, Higgins JP, et al. Seven haemostatic gene polymorphisms in coronary disease: meta-analysis of 66,155 cases and 91,307 controls. Lancet 2006;367:651-8. [Crossref] [PubMed]
- Reitsma PH, Rosendaal FR. Past and future of genetic research in thrombosis. J Thromb Haemost 2007;5 Suppl 1:264-9. [Crossref] [PubMed]
- Yamada Y. Identification of genetic factors and development of genetic risk diagnosis systems for cardiovascular diseases and stroke. Circ J 2006;70:1240-8. [Crossref] [PubMed]
- Topol EJ, McCarthy J, Gabriel S, et al. Single nucleotide polymorphisms in multiple novel thrombospondin genes may be associated with familial premature myocardial infarction. Circulation 2001;104:2641-4. [Crossref] [PubMed]
- Yamada Y, Izawa H, Ichihara S, et al. Prediction of the risk of myocardial infarction from polymorphisms in candidate genes. N Engl J Med 2002;347:1916-23. [Crossref] [PubMed]
- McCarthy JJ, Parker A, Salem R, et al. Large scale association analysis for identification of genes underlying premature coronary heart disease: cumulative perspective from analysis of 111 candidate genes. J Med Genet 2004;41:334-41. [Crossref] [PubMed]
- IBC 50K CAD Consortium , et al. Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet 2011;7:e1002260. [Crossref] [PubMed]
- Roberts R. A genetic basis for coronary artery disease. Trends Cardiovasc Med 2015;25:171-8. [Crossref] [PubMed]
- Prins BP, Lagou V, Asselbergs FW, et al. Genetics of coronary artery disease: genome-wide association studies and beyond. Atherosclerosis 2012;225:1-10. [Crossref] [PubMed]
- International HapMap Consortium. The International HapMap Project. Nature 2003;426:789-96. [Crossref] [PubMed]
- Ozaki K, Ohnishi Y, Iida A, et al. Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction. Nat Genet 2002;32:650-4. [Crossref] [PubMed]
- Schreyer SA, Vick CM, LeBoeuf RC. Loss of lymphotoxin-alpha but not tumor necrosis factor-alpha reduces atherosclerosis in mice. J Biol Chem 2002;277:12364-8. [Crossref] [PubMed]
- McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science 2007;316:1488-91. [Crossref] [PubMed]
- Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007;316:1491-3. [Crossref] [PubMed]
- Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med 2007;357:443-53. [Crossref] [PubMed]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661-78. [Crossref] [PubMed]
- Larson MG, Atwood LD, Benjamin EJ, et al. Framingham Heart Study 100K project: genome-wide associations for cardiovascular disease outcomes. BMC Med Genet 2007;8 Suppl 1:S5. [Crossref] [PubMed]
- Broadbent HM, Peden JF, Lorkowski S, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet 2008;17:806-14. [Crossref] [PubMed]
- Samani NJ, Schunkert H. Chromosome 9p21 and cardiovascular disease: the story unfolds. Circ Cardiovasc Genet 2008;1:81-4. [Crossref] [PubMed]
- Schunkert H, König IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011;43:333-8. [Crossref] [PubMed]
- Reilly MP, Li M, He J, et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet 2011;377:383-92. [Crossref] [PubMed]
- Franchini M, Mannucci PM. ABO blood group and thrombotic vascular disease. Thromb Haemost 2014;112:1103-9. [Crossref] [PubMed]
- Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 2014;42:D1001-6. [Crossref] [PubMed]
- Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015;47:1121-30. [Crossref] [PubMed]
- Myocardial Infarction Genetics Consortium, Kathiresan S, Voight BF, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet 2009;41:334-41. [Crossref] [PubMed]
- Erdmann J, Grosshennig A, Braund PS, et al. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet 2009;41:280-2. [Crossref] [PubMed]
- Davies RW, Wells GA, Stewart AF, et al. A genome-wide association study for coronary artery disease identifies a novel susceptibility locus in the major histocompatibility complex. Circ Cardiovasc Genet 2012;5:217-25. [Crossref] [PubMed]
- Clarke R, Peden JF, Hopewell JC, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med 2009;361:2518-28. [Crossref] [PubMed]
- Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet 2011;43:339-44. [Crossref] [PubMed]
- Willer CJ, Sanna S, Jackson AU, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 2008;40:161-9. [Crossref] [PubMed]


