
MiR-20a: a mechanosensitive microRNA that regulates fluid shear stress-mediated osteogenic differentiation via the BMP2 signaling pathway by targeting BAMBI and SMAD6
Introduction
Mechanical stimulation is critical for both skeletal development as well as bone remodeling. Mechanical stress-mediated deformation of the mineralized matrix leads to heterogeneous pressure gradients that enhance interstitial fluid flow in the Haversian and lacunar-canalicular network systems, generating fluid shear stress (FSS) across bone cell surfaces (1). FSS is widely used for mechanical stimulation of the bone and is a major force involved in bone adaptation. Mechanosensitive cells sense mechanical signals produced by FSS in bone and translate them into biochemical signals (2). Appropriate FSS has a vital function in modulating the functions of bone cells [including osteoblasts, osteocytes, and human adipose-derived stem cells (ASCs)], enhancing cell proliferation, and promoting osteogenic differentiations (3-5). Our previous studies indicated that steady FSS of 12 dyn/cm2 for 1 h promoted the osteogenic differentiation of murine calvaria-derived pre-osteoblasts (MC3T3-E1 cells), human periodontal ligament cells (hPDLCs), and mouse bone marrow stromal cells (BMSCs) (6,7). In addition, biomaterials play an important role in FSS-induced osteogenic differentiation. Pulsatile fluid flow (PFF) aligned on nanostructural biomaterial Increased collagen and alkaline phosphatase mRNA expression in MC3T3-E1 cells (8). FSS in conjunction with Scaffold biomaterials can promote osteogenic differentiation by upregulating alkaline phosphatase (ALP) and Runt-related transcription factor 2 (RUNX2) expression in equine adipose-derived MSC (9).
Mechanotransduction of FSS in mechanically sensitive cells is complicated. Detecting mechanical stimulus is the primary step of mechanotransduction. Mechanical stresses are sensed by different cellular structures, including integrins, integrin-associated kinase, connexins, glycocalyx, and Cytoskeleton (10). Through above cellular structures, mechanical signals can be transmitted to apical structures of cells such as the actin cortical web and the plasma membrane, where signals are transmitted to the intracellular regions (11). Subsequently, mechanosensitive signaling pathways are activated, including Ca2+ signaling pathway, MAPK signaling pathway, Wnt/β-catenin signaling pathway, BMP2 signaling pathway, which translate mechanical stimuli into biochemical signals to regulate the growth, migration, and differentiation of cell (12).
Bone morphogenetic protein 2 (BMP2) is a key regulatory factor of osteoblast differentiation. Kamiya et al. demonstrated that the BMP2 signaling pathway plays a vital role in the transcriptional control of bone formation (13). The BMP2 intracellular receptor mothers against decapentaplegic family members (SMADs), especially SMAD5, activate the transcription of osteogenic-associated genes, such as RUNX2/Cbfa-1 and SP7 transcription factor (SP7)/Osterix, which in turn leads to extracellular matrix (ECM) mineralization to generate osteoids (14). It has been reported that mechanical strain increases the phosphorylation of SMAD1/5 and activates the BMP2/SMAD pathway in osteoblast differentiation (15,16). Moreover, Liu et al. showed that FSS promotes BMP2 secretion and activates the BMP2 signaling pathway, highlighting the crucial function of the BMP2 signaling pathway in mechanical loading-induced osteogenic differentiation (17). Similarly, our previous results showed that the association between the BMP2 and integrin β1 pathways plays a vital role in FSS-induced osteogenic differentiation. However, the underlying mechanisms require further investigation.
MicroRNAs (miRNAs), which form a class of non-coding small (18 to 25 nucleotides) RNAs, are key negative regulators of various biological and pathological processes, including apoptosis, development, organogenesis, cell proliferation, and differentiation (18,19). Mature miRNAs post-transcriptionally suppress gene expression levels by binding their regulatory targets in 3'-untranslated regions (3'-UTRs) of messenger RNAs (mRNAs), which leads to mRNA degradation or translational suppression (18,20). Various miRNAs play significant roles in the regulation of chemical stimulation-mediated osteogenic differentiation as well as bone formation in vitro. For example, miR-20a, miR-10a, miR-96, and miR-21 promote osteogenic differentiation (21-24), while miR-200a, miR-138, miR-141, and miR-133a-5p inhibit osteogenic differentiation (25-27). Thus, it has been strongly postulated that the regulation of osteogenic differentiation by miRNAs is an important aspect of the regulatory machinery. Furthermore, several studies have evaluated the mechanical force-mediated changes in the expressions of miRNAs associated with bone metabolism. For example, miR-103-a is suppressed during cyclical mechanical stretching-mediated osteoblast differentiation by targeting pank3, leading to elevated RUNX2 protein levels (28). Additionally, miR-503-5p functions as a mechanosensitive miRNA and suppresses the osteogenic differentiation of BMSCs in mechanically-stretched bones and bone formation in orthodontic teeth movement (OTM) tension sides (29). The exposure of PDLSCs to a stretching force suppresses activin receptor type IIB (ACVR2B) levels via direct interactions between miR-21 and 3-untranslated repeat sequence of the ACVR2B mRNA (30). Moreover, FSS-induced osteoblast differentiation is negatively regulated by miR-33-5p as well as its target gene (Hmga2) through the regulation of stem cells and MSC proliferation (31). These findings imply that miRNAs play vital roles in the sensing of mechanical loads by osteoblasts. Nevertheless, the impacts of FSS mechanical stimuli on miRNA expression as well as their roles in mechanotransduction in osteoblasts have not yet been fully elucidated. Therefore, further studies are required to confirm the associations between miRNA expression levels and mechanical force. To determine the functions of miRNAs in the mechanotransduction of FSS, we screened for mechanosensitive miRNAs using the miRNA microarray strategy and found miR-20a to be distinctly changed in MC3T3-E1 cells in response to FSS (32).
We identified differentially-expressed miRNAs that are putatively related to the BMP2 signaling pathway and performed a miR-20a spatial-temporal up-regulation pattern during FSS-mediated osteogenic differentiation of MC3T3-E1 pre-osteoblastic cells or mouse BMSCs. MiR-20a was found to be a positive regulator of FSS-induced osteogenic differentiation through activation of the BMP2 signaling pathway via direct down-regulation of BAMBI and SMAD6, which are negative factors of the BMP2 signaling pathway. These results imply that miR-20a is a mechanosensitive microRNA which has a significant function in FSS-induced osteogenic differentiation. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2753/rc).
Methods
Cell culture
Culturing of MC3T3-E1 cells, a murine pre-osteoblastic cell line (ATCC, Manassas, VA, USA) was performed in minimum essential medium alpha medium (α-MEM) media (Life Technologies, Grand Island, NY, USA) with 1% penicillin/streptomycin (Life Technologies) and fetal bovine serum (FBS; 10%, Life Technologies). Incubation was carried out in a 5% CO2 humidified environment at 37 ℃. The medium was replaced every 3 days. The mouse BMSCs were isolated as described previously (33) and preserved in DMEM with 10% FBS. The multipotential differentiation ability was then determined (Figure S1).
Application of fluid flow stress
After the cells grew to a confluence of 80–85% on polylysine (10 µg/mL; Sigma-Aldrich, USA) coated glass slides, cell starvation was performed for 12 h in a serum-free media. Next, the cells were treated with FSS (12 dyn/cm2) for 1 h using a parallel plate flow system that generated a laminar unidirectional flow across the cells as previously described (6). Flow system maintenance was conducted at 37 ℃ after being filled with a 5% CO2 serum-supplemented medium (10%).
The control cells were cultured at a static station. To conduct the control experiments, the cells were placed in identical conditions as those of equivalent FSS experiments, but without FSS stimulus application. Following FSS stimulation, cell growth was performed under identical osteogenic conditions [β-glycerophosphate (10 mM) and ascorbic acid (50 µg/mL) for MC3T3-E1; β-glycerophosphate (10 mM), ascorbic acid (50 µg/mL), and dexamethasone (10 nM) for mouse BMSCs].
RNA isolation, reverse transcription, and real-time quantitative polymerase chain reaction (qRT-PCR)
Trizol reagent (Life Technologies) was used for total RNA extraction. The expression levels of mature miRNA were evaluated using stem-loop primer SYBR Green qRT-PCR and were normalized to U6. The gene transcriptional levels were evaluated by qRT-PCR. Normalization of the expression levels was conducted to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. QRT-PCR was performed using a 7500 Real-Time PCR system (Applied Biosystems, USA) using Platinum SYBR Green qPCR SuperMix-UDG (Life Technologies). The mRNA expression levels were calculated by the CT method (△△CT) using GAPDH as an internal reference. The qRT-PCR procedure comprised 40 cycles (15 s at 94 ℃, 60.5 ℃ for 15 s, 15 s at 72 ℃), which followed an initial denaturation step (2 min at 94 ℃). The primers for this experiment are presented in Table S1.
Immunoblotting
Quantification was performed using a bicinchoninic acid (BCA) protein assay kit (Pierce, Rockford, IL, USA). SDS-PAGE (12%) was then used to separate the cell lysates, followed by their transfer to PVDF membranes. Membrane blocking was conducted using non-fat dry milk (5%) for 1 h followed by overnight incubation in the presence of primary antibodies against BAMBI (1:1,000; R&D, USA), SMAD6 (1:500; AVIVA, USA), PPARγ (1:1,000; Cell Signaling Technology, USA), BMP2 (1:400; Bioss Company, China), or RUNX2 (1:400; Bioss Company, China). After washing, the membranes were incubated with an HRP-conjugated secondary antibody (Santa Cruz Biotechnology, USA). Detection was performed by chemiluminescence. The GAPDH protein was the internal loading control. The intensities of the protein fragments were quantified using Image Lab software version 4.0 (BIO-RAD, USA).
ALP activity assessment
Cellular ALP activities were assessed using the nitrophenyl phosphate (PNPP) method. P-nitrophenylphosphate was used as the substrate (LabAssay TM ALP, Wako, Japan). Enzymatic activities (units/mg protein) were equal to p-nitrophenol (nmol/mL) concentrations released by the sample within 15 min, after background exclusion. The ALP activities of every sample were normalized to protein concentrations measured using a BCA protein assay kit (Thermo Scientific Pierce, USA).
Alizarin red S (ARS) staining
To evaluate ECM mineralization as a terminal marker of differentiation, the cells were washed, fixed in paraformaldehyde (4%), and stained using ARS solution (1%; Sigma-Aldrich). Imaging was conducted randomly by light microscopy (400×, Olympus). Next, ARS stain quantification was performed by extraction using cetyl-pyridinium chloride monohydrate (TCI, Japan). The measurement of absorbance was carried out at 560 nm.
Oligonucleotide transfection
Transfection of miR-20a mimics, miR-20a inhibitor (anti-miR-20a), negative control (NC), BAMBI silencing oligonucleotides (si-BAMBI), inhibitor negative control (inhibitor-NC), and SMAD6 silencing oligonucleotides (si-SMAD6) (RiboBio, China) into cells was performed using Lipofectamine RNAiMAX (Invitrogen, USA) according to the manufacturer’s instructions. The efficiency of transfection was assessed using Cy3-labeled oligonucleotide RNA mimics (Figure S2).
Luciferase assay
Possible binding sites for miR-20a within BAMBI or SMAD6 3'-UTR were determined using PicTar (http://pictar.mdc-berlin.de/) and TargetScan (www.targetscan.org). Annealed synthetic oligonucleotides and predicted binding sites were cloned into the XhoI/NotI site of psiCHECK-2 (Promega, USA). Transient transfection of Cos-7 cells with a mutant (MUT) reporter vector or wild-type (WT) and microRNA was performed using Lipofectamine 2000 (Invitrogen) at various concentrations. At 48-hour post-transfection, luciferase activity was assessed using the Dual-Luciferase Reporter Assay System (Promega, USA) according to the manufacturer’s instructions. Normalization of the Renilla luciferase activities was conducted to the equivalent firefly luciferase activities and plotted as percentages of the control.
RNA interference (RNAi)
The design and synthesis of double-stranded small interfering RNA (siRNAs) were performed using RiboBio (Guangzhou, China). Transfection of siRNAs (50 nM) was conducted using Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer’s protocol. Double-stranded scrambled RNA was the NC. Transfection of cells that had been seeded on glass slides was carried out for 12 h, after which they were subjected to additional treatments.
Statistical analysis
Data are presented as mean ± SD for n=3. Comparisons were made by using a two-tailed t-test or one-way ANOVA for experiments with more than two subgroups. Statistical significance was attained at P<0.05 (*, P<0.05; **, P<0.01; ***, P<0.001.).
Results
MiR-20a is differentially up-regulated during steady FSS-mediated osteogenic differentiation
Osteogenic differentiation in MC3T3-E1 pre-osteoblastic cells was initiated by a 12 dyn/cm2 single load of FSS for 1 h and confirmed by elevated expression levels of osteogenic differentiation biomarker genes, including RUNX2, ALP, and SP7 at 12 h after FSS treatment (pf.) (Figure 1A). Confirmation of the osteoblast phenotypes was conducted using the detection of enhanced ALP activities (Figure 1B) and ARS staining for ECM mineralization (Figure 1C). The elevated expression levels of osteoblast-related genes as well as the observed osteoblast phenotypes were consistent with the previously reported observations of FSS-promoted osteogenic MC3T3-E1 cell differentiation.
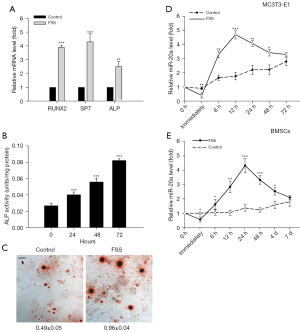
To identify candidate miRNAs that regulate FSS-induced osteogenic differentiation, miRNA array profiling was conducted after treating MC3T3-E1 cells with FSS (12 dyn/cm2) using Agilent miRNA chips (SBC, China). There were significant changes in expression levels of 69 miRNAs after FSS-treatment, which was performed as described previously (32). Functional analysis of these miRNAs revealed that they were closely related to the BMP2 signaling pathway, in which 11 miRNAs are involved (Table S2). QRT-PCR analysis revealed the spatiotemporal expression pattern of miR-20a, -19b, and -34a of 11 miRNAs during FSS-induced osteogenic MC3T3-E1 differentiation, which were reduced immediately for a short time, but quickly elevated at 6 h, then peaked at 12 h, and remained elevated relative to the control until 72 h (Figure 1D, Figure S3). We used primary mouse BMSCs to confirm this outcome. Similarly, the expression of miR-20a decreased immediately for a short period, but consistently increased at 6 h post-FSS treatment, then peaked at 24 h, and remained higher than the control until day 7 (Figure 1E), indicating that miR-20a was an early response gene for FSS in osteoblasts.
Overexpression of miR-20a activates chemical stimulation-mediated osteogenic MC3T3-E1 cell differentiation
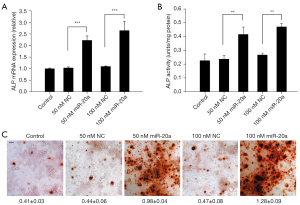
Overexpression studies were performed to determine the biological functions of miR-20a in osteogenic differentiation (Figure 2). Overexpressed miR-20a elevated the mRNA expression of ALP at 48 h and increased ALP activity on day 3 (Figure 2A,2B). An ARS staining experiment was conducted to ascertain the osteogenic effects of miR-20a. Figure 2C showed that overexpressed miR-20a enhanced matrix mineralization on day 14. Therefore, overexpression of miR-20a increases osteogenic MC3T3-E1 cell differentiation induced by the standard osteoblast-induction medium.
MiR-20a promotes FSS-mediated osteogenic differentiation by promoting the BMP2 signaling pathway
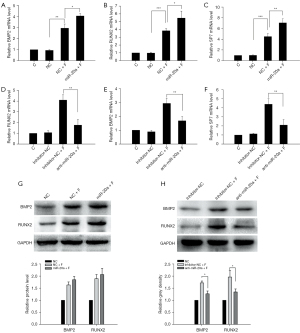
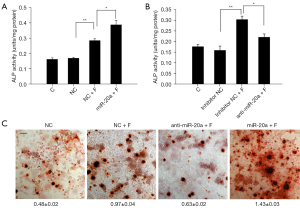
Previous reports have indicated that the canonical BMP2 signaling pathway plays crucial roles in FSS-mediated osteogenic differentiation. The function of miR-20a in FSS-mediated osteogenic differentiation was further determined by investigating the effects of the loss (anti-miR-20a) or overexpression of miR-20a on osteogenic MC3T3-E1 cell differentiation in vitro. After FSS treatment (pf.), overexpression of miR-20a significantly increased the transcriptional levels of RUNX2, BMP2, and SP7, which are the main regulators of the BMP2/RUNX2 pathway (Figure 3A-3C). However, a notable decrease in the transcriptional levels of BMP2, RUNX2, and SP7 was observed in the anti-miR-20a group after FSS treatment (pf.), although this was still higher than the without FSS-treatment groups (Figure 3D-3F). Accordingly, the BMP2 and RUNX2 protein levels were suppressed in the anti-miR-20a group, although overexpression of miR-20a only slightly increased their protein levels during FSS-induced osteogenic differentiation (Figure 3G,3H). Furthermore, we observed enhanced ALP activity with overexpression of miR-20a (Figure 4A), which were attenuated remarkably with anti-miR-20a during FSS-induced osteogenic differentiation (Figure 4B). Similarly results were observed for nodule formation during FSS-induced osteogenic differentiation (Figure 4C). These results suggest that up-regulated miR-20a promotes FSS-induced osteogenic differentiation and bone formation by activating the BMP2 signaling pathway.
MiR-20a activates the BMP2 signaling pathway by directly targeting BAMBI and SMAD6
To understand the functional roles of miR-20a during FSS-promoted osteogenic differentiation, the microRNA target prediction software, TargetScan and PicTar, were used to search for potential miR-20a targets (Table S3). BAMBI, SMAD6, and PPARγ, which are antagonists of the BMP2/RUNX2 signaling pathway, were identified as putative target miR-20a genes. To ascertain whether they are potential miR-20a targets, we analyzed the expressions of BAMBI, SMAD6, and PPARγ pf. In both the FSS-treatment and miR-20a overexpression groups, there was a marked decrease in BAMBI and SMAD6 protein levels in the MC3T3-E1 cells (Figure 5A). However, alterations in the mRNA levels of BAMBI and SMAD6 were not significant pf. (Figure 5B), indicating that the effect was post-transcriptional. We also confirmed our findings using primary mouse BMSCs. Consistent with the findings in MC3T3-E1 cells, we observed significantly low protein levels of BAMBI and SMAD6 but not mRNA in both the FSS-treatment and miR-20a overexpression groups (Figure 5C,5D). However, no significant changes in the protein or mRNA levels of PPARγ were observed both in MC3T3-E1 cells and in primary mouse BMSCs (Figure S4).
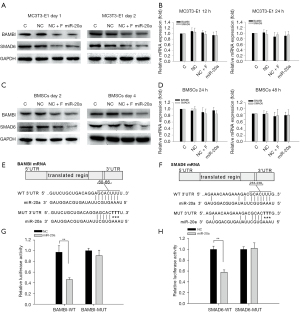
According to the TargetScan bioinformatics prediction, BAMBI and SMAD6 have a 7-nucleotide seed match site for miR-20a within their 3'-UTR, and the putative target site is highly conserved in vertebrate species (Figure 5E,5F). To determine whether miR-20a can directly target BAMBI and SMAD6, the 3'-UTR of BAMBI or SMAD6 was amplified, and the luciferase reporter vector psiCHECK2 was then inserted into the 3'-UTR. Reporter vectors exhibiting mutated putative target sites were also inserted. The mutated sites of the 3'UTR of BAMBI or SMAD6 are shown in Figure 5E,5F. Ectopic miR-20a was co-transfected with BAMBI-WT, SMAD6-WT, BAMBI-MUT, or SMAD6-MUT vectors into cos-7 cells and luciferase activity was subsequently assessed. Compared to the NC, the luciferase activity of the BAMBI-WT sequence was decreased by 47.7% through miR-20a. However, luciferase activities were not affected by miR-20a in the BAMBI-MUT sequence (Figure 5G). Similarly, relative to NC, the luciferase activity of the SMAD6-WT sequence was decreased by 40.3% through miR-20a and was not affected by miR-20a in the SMAD6-MUT sequence (Figure 5H). In summary, BAMBI and SMAD6 are direct targets for miR-20a.
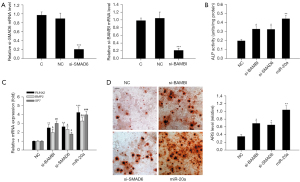
Silencing of BAMBI and SMAD6 mimics the osteogenic effects of miR-20a
To determine whether BAMBI and SMAD6 are functional targets of miR-20a during osteogenic differentiation, we further determined whether the inhibition of BAMBI and SMAD6 expressions mimicked the differentiation-enhancing effects of miR-20a. Figure 6A shows that the mRNA expression levels of BAMBI and SMAD6 were effectively repressed in MC3T3-E1 cells with RNAi for 24 h. Silencing of BAMBI and SMAD6 enhanced ALP activity (Figure 6B). Furthermore, silencing of BAMBI and SMAD6 resulted in increased transcription of osteoblast-associated genes (BMP2, RUNX2, and ALP) (Figure 6C) as well as ECM mineralization (Figure 6D). We noticed that the osteogenic effects of BAMBI and SMAD6 knockdown were weaker than those of miR-20a overexpression, indicating that miR-20a may target other genes as well. These results indicate that knockdown of BAMBI or SMAD6 mimics the osteogenic differentiation of miR-20a.
Discussion
Mechanical force-induced osteoblast differentiation is a basic feature of bone homeostasis, such as orthodontic tooth movement and bone repair. MiRNAs respond to mechanical loading and are involved in osteoblast differentiation as well as bone formation (34). We found that miR-20a is a novel mechanosensitive miRNA that is involved in the positive regulation of osteoblast differentiation via activation of the BMP2 signaling pathway by repressing the expression of BAMBI and SMAD6 post-transcriptionally. Thus, miR-20a is a possible therapeutic target for pathological disorders that are associated with skeletal development induced by mechanical environment changes.
FSS, a potent form of bone mechanical stimulation, plays a vital role in the differentiation, function, and development of osteoblasts. Important molecules and pathways involved in FSS-induced osteoblast differentiation have been identified. For example, FSS activates extracellular signal-regulated kinase 1/2 (ERK1/2), beta-catenin (β-catenin), prostaglandin E2 (PGE2), and glycogen synthase kinase-3β (GSK-3β) which are vital signaling constituents of mechanotransduction pathways in osteoblasts that enhance cellular differentiation (35-37). It has been reported that several signaling pathways, including Wnt/β-catenin, Ca2+, and BMP2 signaling, play various roles in FSS-mediated osteoblast differentiation, through which biochemical process are triggered (35,36,38). In addition, we previously discovered that the association between the integrinβ1 and BMP2 pathways plays a vital role in these differentiations (6).
Recent studies have demonstrated the mechanosensitive properties of miRNAs in cells under FSS. For example, the up-regulation of miR-132 plays a role in FSS-induced differentiation and proliferation in periodontal ligament cells (39). Similarly, miR-33-5P is elevated and promotes osteoblast MC3T3-E1 cell differentiation under FSS (31). FSS results in the up-regulation of E-Tmod41 via miR-23b-3p inhibition as well as PE0 promoter activation, thereby contributing to F-actin cytoskeletal remodeling (40). In rat hind limbs, the down-regulation of miR-352 is involved in the elevation of FSS-mediated collateral vessel growth (41). The oscillatory shear-sensitive miR-181b regulates aortic valve endothelial matrix degradation (42). We found that miR-20a expression increased significantly 6 h after FSS in MC3T3-E1 cells (12 h in BMSCs). This is the first study to report a miRNA that is involved in activating the BMP2 signaling pathway by FSS. According in response to FSS, we established a spatiotemporal expression pattern of miR-20a in MC3T3-E1 cells or BMSCs, and found that miR-20a, as a mechanosensitive miRNA, is sensitive to FSS.
Among the miRNA families, miR-20a, which is a miR-17-92 cluster member, has been widely evaluated. This family has vital functions in both tissue and organ development, and is closely related to tumor development (43), autoimmune diseases (26), and osteogenesis (44). MiR-20a is expressed in mice and conserved in humans, and is especially elevated in osteoblasts and bone tissues (45). It was reported that the miR-17-92 cluster can suppress transforming growth factor beta (TGFB) pathway-mediated proliferation inhibition and collagen synthesis in palatal mesenchymal cells (PMCs) by targeting SMAD4, TGFBR2, and SMAD2 (46). MiR-20a also plays a role in the osteogenic differentiation of human mesenchymal stem cells (MSCs) via the BMP2/RUNX2 signaling pathway by targeting Crim1, PPARγ, and BAMBI (21). In addition, up-regulation of miR-20a promotes the osteogenic differentiation of human adipose tissue-derived stem cells (hASCs) (47). Moreover, a recent study showed that miR-20a negatively regulates THP-1 human acute monocytic leukemia cells proliferation and osteoclastogenesis during osteoclast differentiation by down-regulating PPARγ (48). These studies imply that miR-20a plays a vital role in the regulation of osteogenic differentiation and osteogenesis. Nevertheless, the significance of miR-20a in mechanotransduction has not been established. Our findings also demonstrate that miR-20a is involved in the regulation of osteoblast differentiations in mouse BMSCs and MC3T3-E1 cells. Our results showed that overexpressed miR-20a elevates the expression levels of canonical markers associated with osteoblast differentiation, such as RUNX2 and SP7. Furthermore, the ability of miR-20a to enhance osteoblast differentiations were confirmed in simulated FSS environments. Overexpressed miR-20a promoted osteoblast differentiation induced by simulated FSS. Accordingly, miR-20a knockdown inhibited this differentiation, implying that miR-20a participates in the modulation of FSS-induced osteoblast differentiation.
The BMP2 signaling pathway plays a vital role in the transcriptional regulation of bone formation. Our data demonstrated that miR-20a promotes FSS-mediated osteogenic differentiation by promoting the BMP2 signaling pathway. BMP2 and RUNX2 were up-regulated at both the mRNA and protein levels. However, their associations may not be direct since miRNAs regulate gene expressions by binding the 3'-UTRs of target genes, which leads to translational suppression or mRNA degradation. Consequently, miR-20a may activate the BMP2 signaling pathway by inhibiting its antagonists or negative regulators.
MiRNAs have hundreds of target sites and regulate a majority of protein-coding genes, thereby forming a regulatory network (49). Based on our findings, SMAD6 and BAMBI possess high-score miR-20a binding sites in their 3'-UTRs, indicating that they may be targets for miR-20a in the BMP2 signaling pathways. BAMBI is a BMP2 signaling pathway pseudo-receptor whose amino acid sequence is highly to comparable that of BMP type-I receptors, BMPR-IB and BMPR-IA. It has been shown that BAMBI inactivates ligand-receptor complexes and antagonizes BMP signaling by associating with BMP ligands and functional BMP receptors (50). SMAD6, a SMAD family member, functions as a suppressor of certain SMAD family members. SMAD6 suppresses SMAD1 phosphorylation under the induction of the bone morphogenetic protein type IB receptor. SMAD6 competes with SMAD4 to bind the receptor-activated SMAD1/5/8 complex to form an inactive R-SMAD polymer, which interferes with BMP2/SMADs signal transduction (51). In addition, SMAD6 promotes SmurF1-mediated RUNX2 degradation, thereby inhibiting BMP2/RUNX2 signaling pathway activation (52).
In this study, both BAMBI and SMAD6 were determined to be regulators of miR-20a that participate in the activation of the BMP2/RUNX2 signaling pathway, which regulates osteoblast differentiation in FSS-simulated environments. We observed that the protein expressions of BAMBI and SMAD6 were suppressed during FSS-induced osteogenic differentiation, similar to the overexpression of miR-20a. We postulated that this decrease of the negative regulator of the BMP2/RUNX2 signaling pathway might activate BMP2 signaling, thereby enhancing osteogenic differentiation. These results suggest a miR-20a co-regulatory mechanism in osteoblast differentiation, as was demonstrated in our study (Figure 7).
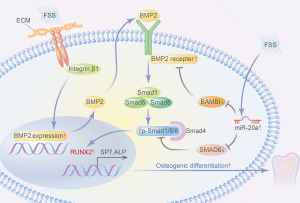
Zhang et al. reported that miR-20a plays a key role in hMSC osteogenic differentiation. This effect is achieved through the BMP/RUNX2 signaling pathway by targeting PPARγ, BAMBI, and Crim1 (21). We also confirmed that miR-20a regulated FSS-induced osteogenic differentiations of MC3T3-E1 cells and mouse BMSCs via the BMP2 signaling pathway by targeting BAMBI and SMAD6. However, PPARγ did not appear to be directly targeted by miR-20a. These findings indicate that the functions of miR-20a are highly conserved and similar across different species. Conversely, it also implies that miR-20a targets may differ among species.
There are a few limitations of our study that are worth noting. In accordance with previous investigations, we evaluated the effects of 12 dyn/cm2 single bout FSS (for 1 hour) on osteoblast differentiations. However, the magnitude and application time of FSS may have had an effect on osteoblast differentiation. Therefore, the significance of miR-20a in FSS-mediated osteoblast differentiation should be further evaluated in various FSS environments. Furthermore, our in vitro findings were not confirmed in vivo.
Conclusions
In summary, as a particular mechanosensitive miRNA, miR-20a senses the mechanical environment in osteoblasts and moderates their differentiation. Specifically, miR-20a suppresses several BMP2 signaling pathway-associated genes at post-transcriptional levels to yield mild regulations in individual target genes and thereby ultimately enhance osteoblast MC3T3-E1 cell osteogenesis. Our results elucidated the mechanisms of mechanotransduction in osteoblasts and implied that miRNAs may be targeted in bone tissue engineering and orthodontic bone remodeling for regenerative medicine applications.
Acknowledgments
We thank Professor Guangmei Yan (Sun Yat-sen University) and his group for providing technical support.
Funding: This work was supported by grants from the National Natural Science Foundation of China (No. 81470766), the Natural Science Foundation of Guangdong Province (No. 81070860) and Projects of General Program By the Natural Science Foundation of Guangdong Province (No. 2016A030313212).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-2753/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-2753/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2753/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fritton SP, Weinbaum S. Fluid and Solute Transport in Bone: Flow-Induced Mechanotransduction. Annu Rev Fluid Mech 2009;41:347-74. [Crossref] [PubMed]
- Li Y, Wang J, Xing J, et al. Surface chemistry regulates the sensitivity and tolerability of osteoblasts to various magnitudes of fluid shear stress. J Biomed Mater Res A 2016;104:2978-91. [Crossref] [PubMed]
- Aisha MD, Nor-Ashikin MN, Sharaniza AB, et al. Orbital fluid shear stress promotes osteoblast metabolism, proliferation and alkaline phosphates activity in vitro. Exp Cell Res 2015;337:87-93. [Crossref] [PubMed]
- Zhang B, An L, Geng B, et al. ERK5 negatively regulates Kruppel-like factor 4 and promotes osteogenic lineage cell proliferation in response to MEK5 overexpression or fluid shear stress. Connect Tissue Res 2021;62:194-205. [Crossref] [PubMed]
- Schneider I, Baumgartner W, Gröninger O, et al. 3D microtissue-derived human stem cells seeded on electrospun nanocomposites under shear stress: Modulation of gene expression. J Mech Behav Biomed Mater 2020;102:103481. [Crossref] [PubMed]
- Mai Z, Peng Z, Wu S, et al. Single bout short duration fluid shear stress induces osteogenic differentiation of MC3T3-E1 cells via integrin β1 and BMP2 signaling cross-talk. PLoS One 2013;8:e61600. [Crossref] [PubMed]
- Tang M, Peng Z, Mai Z, et al. Fluid shear stress stimulates osteogenic differentiation of human periodontal ligament cells via the extracellular signal-regulated kinase 1/2 and p38 mitogen-activated protein kinase signaling pathways. J Periodontol 2014;85:1806-13. [Crossref] [PubMed]
- Prodanov L, Semeins CM, van Loon JJ, et al. Influence of nanostructural environment and fluid flow on osteoblast-like cell behavior: a model for cell-mechanics studies. Acta Biomater 2013;9:6653-62. [Crossref] [PubMed]
- Elashry MI, Baulig N, Wagner AS, et al. Combined macromolecule biomaterials together with fluid shear stress promote the osteogenic differentiation capacity of equine adipose-derived mesenchymal stem cells. Stem Cell Res Ther 2021;12:116. [Crossref] [PubMed]
- Liu L, Yuan W, Wang J. Mechanisms for osteogenic differentiation of human mesenchymal stem cells induced by fluid shear stress. Biomech Model Mechanobiol 2010;9:659-70. [Crossref] [PubMed]
- Tarbell JM, Weinbaum S, Kamm RD. Cellular fluid mechanics and mechanotransduction. Ann Biomed Eng 2005;33:1719-23. [Crossref] [PubMed]
- Wang L, You X, Zhang L, et al. Mechanical regulation of bone remodeling. Bone Res 2022;10:16. [Crossref] [PubMed]
- Kamiya N, Mishina Y. New insights on the roles of BMP signaling in bone-A review of recent mouse genetic studies. Biofactors 2011;37:75-82. [Crossref] [PubMed]
- Matsubara T, Kida K, Yamaguchi A, et al. BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J Biol Chem 2008;283:29119-25. [Crossref] [PubMed]
- Rath B, Nam J, Deschner J, et al. Biomechanical forces exert anabolic effects on osteoblasts by activation of SMAD 1/5/8 through type 1 BMP receptor. Biorheology 2011;48:37-48. [Crossref] [PubMed]
- Wang L, Zhang X, Guo Y, et al. Involvement of BMPs/Smad signaling pathway in mechanical response in osteoblasts. Cell Physiol Biochem 2010;26:1093-102. [Crossref] [PubMed]
- Liu L, Shao L, Li B, et al. Extracellular signal-regulated kinase1/2 activated by fluid shear stress promotes osteogenic differentiation of human bone marrow-derived mesenchymal stem cells through novel signaling pathways. Int J Biochem Cell Biol 2011;43:1591-601. [Crossref] [PubMed]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15-20. [Crossref] [PubMed]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 2011;39:D152-7. [Crossref] [PubMed]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5' UTR as in the 3' UTR. Proc Natl Acad Sci U S A 2007;104:9667-72. [Crossref] [PubMed]
- Zhang JF, Fu WM, He ML, et al. MiRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biol 2011;8:829-38. [Crossref] [PubMed]
- Sun Y, Xu L, Huang S, et al. mir-21 overexpressing mesenchymal stem cells accelerate fracture healing in a rat closed femur fracture model. Biomed Res Int 2015;2015:412327. [Crossref] [PubMed]
- Dong J, Zhang Z, Huang H, et al. miR-10a rejuvenates aged human mesenchymal stem cells and improves heart function after myocardial infarction through KLF4. Stem Cell Res Ther 2018;9:151. [Crossref] [PubMed]
- Yang M, Pan Y, Zhou Y. miR-96 promotes osteogenic differentiation by suppressing HBEGF-EGFR signaling in osteoblastic cells. FEBS Lett 2014;588:4761-8. [Crossref] [PubMed]
- Sangani R, Periyasamy-Thandavan S, Kolhe R, et al. MicroRNAs-141 and 200a regulate the SVCT2 transporter in bone marrow stromal cells. Mol Cell Endocrinol 2015;410:19-26. [Crossref] [PubMed]
- Eskildsen T, Taipaleenmäki H, Stenvang J, et al. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci U S A 2011;108:6139-44. [Crossref] [PubMed]
- Zhang W, Wu Y, Shiozaki Y, et al. miRNA-133a-5p Inhibits the Expression of Osteoblast Differentiation-Associated Markers by Targeting the 3' UTR of RUNX2. DNA Cell Biol 2018;37:199-209. [Crossref] [PubMed]
- Zuo B, Zhu J, Li J, et al. microRNA-103a functions as a mechanosensitive microRNA to inhibit bone formation through targeting Runx2. J Bone Miner Res 2015;30:330-45. [Crossref] [PubMed]
- Liu L, Liu M, Li R, et al. MicroRNA-503-5p inhibits stretch-induced osteogenic differentiation and bone formation. Cell Biol Int 2017;41:112-23. [Crossref] [PubMed]
- Wei F, Liu D, Feng C, et al. microRNA-21 mediates stretch-induced osteogenic differentiation in human periodontal ligament stem cells. Stem Cells Dev 2015;24:312-9. [Crossref] [PubMed]
- Wang H, Sun Z, Wang Y, et al. miR-33-5p, a novel mechano-sensitive microRNA promotes osteoblast differentiation by targeting Hmga2. Sci Rep 2016;6:23170. [Crossref] [PubMed]
- Mai ZH, Peng ZL, Zhang JL, et al. miRNA expression profile during fluid shear stress-induced osteogenic differentiation in MC3T3-E1 cells. Chin Med J (Engl) 2013;126:1544-50. [PubMed]
- Chen Y, Dong XJ, Zhang GR, et al. In vitro differentiation of mouse bone marrow stromal stem cells into hepatocytes induced by conditioned culture medium of hepatocytes. J Cell Biochem 2007;102:52-63. [Crossref] [PubMed]
- Wang Y, Jia L, Zheng Y, et al. Bone remodeling induced by mechanical forces is regulated by miRNAs. Biosci Rep 2018;38:BSR20180448. [Crossref] [PubMed]
- Norvell SM, Alvarez M, Bidwell JP, et al. Fluid shear stress induces beta-catenin signaling in osteoblasts. Calcif Tissue Int 2004;75:396-404. [Crossref] [PubMed]
- Liu D, Genetos DC, Shao Y, et al. Activation of extracellular-signal regulated kinase (ERK1/2) by fluid shear is Ca(2+)- and ATP-dependent in MC3T3-E1 osteoblasts. Bone 2008;42:644-52. [Crossref] [PubMed]
- Galea GL, Sunters A, Meakin LB, et al. Sost down-regulation by mechanical strain in human osteoblastic cells involves PGE2 signaling via EP4. FEBS Lett 2011;585:2450-4. [Crossref] [PubMed]
- Riddle RC, Donahue HJ. From streaming-potentials to shear stress: 25 years of bone cell mechanotransduction. J Orthop Res 2009;27:143-9. [Crossref] [PubMed]
- Qi L, Zhang Y. The microRNA 132 regulates fluid shear stress-induced differentiation in periodontal ligament cells through mTOR signaling pathway. Cell Physiol Biochem 2014;33:433-45. [Crossref] [PubMed]
- Mu W, Wang X, Zhang X, et al. Fluid Shear Stress Upregulates E-Tmod41 via miR-23b-3p and Contributes to F-Actin Cytoskeleton Remodeling during Erythropoiesis. PLoS One 2015;10:e0136607. [Crossref] [PubMed]
- Guan Y, Cai B, Wu X, et al. microRNA-352 regulates collateral vessel growth induced by elevated fluid shear stress in the rat hind limb. Sci Rep 2017;7:6643. [Crossref] [PubMed]
- Heath JM, Fernandez Esmerats J, Khambouneheuang L, et al. Mechanosensitive microRNA-181b Regulates Aortic Valve Endothelial Matrix Degradation by Targeting TIMP3. Cardiovasc Eng Technol 2018;9:141-50. [Crossref] [PubMed]
- Tipanee J, Di Matteo M, Tulalamba W, et al. Validation of miR-20a as a Tumor Suppressor Gene in Liver Carcinoma Using Hepatocyte-Specific Hyperactive piggyBac Transposons. Mol Ther Nucleic Acids 2020;19:1309-29. [Crossref] [PubMed]
- Li Z, Hassan MQ, Volinia S, et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A 2008;105:13906-11. [Crossref] [PubMed]
- Zhou M, Ma J, Chen S, et al. MicroRNA-17-92 cluster regulates osteoblast proliferation and differentiation. Endocrine 2014;45:302-10. [Crossref] [PubMed]
- Li L, Shi JY, Zhu GQ, et al. MiR-17-92 cluster regulates cell proliferation and collagen synthesis by targeting TGFB pathway in mouse palatal mesenchymal cells. J Cell Biochem 2012;113:1235-44. [Crossref] [PubMed]
- Luo T, Yang X, Sun Y, et al. Effect of MicroRNA-20a on Osteogenic Differentiation of Human Adipose Tissue-Derived Stem Cells. Cells Tissues Organs 2019;208:148-57. [Crossref] [PubMed]
- Wang H, Shen Y. MicroRNA-20a negatively regulates the growth and osteoclastogenesis of THP-1 cells by downregulating PPARγ. Mol Med Rep 2019;20:4271-6. [Crossref] [PubMed]
- Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol 2010;11:252-63. [Crossref] [PubMed]
- Onichtchouk D, Chen YG, Dosch R, et al. Silencing of TGF-beta signalling by the pseudoreceptor BAMBI. Nature 1999;401:480-5. [Crossref] [PubMed]
- Imamura T, Takase M, Nishihara A, et al. Smad6 inhibits signalling by the TGF-beta superfamily. Nature 1997;389:622-6. [Crossref] [PubMed]
- Shen R, Chen M, Wang YJ, et al. Smad6 interacts with Runx2 and mediates Smad ubiquitin regulatory factor 1-induced Runx2 degradation. J Biol Chem 2006;281:3569-76. [Crossref] [PubMed]


