
The synergistic effect of CDKN2B-AS1 and SPC25 on triple-negative breast cancer
Introduction
In 2020, breast cancer (BC) was one of the most common cancers and the primary cause of cancer-related mortality in women worldwide (1). Despite crucial advances in the accurate diagnosis and early intervention of BC, the survival times of some patients remain short. BC is a heterogeneous disease that manifests in different molecular and clinicopathologic features (2). Triple-negative breast cancer (TNBC) is the most aggressive and fatal subtype of BC and accounts for 10–20% of all BC cases (3). Thus, novel therapeutic targets for TNBC desperately need to be identified.
Spindle component 25 (SPC25) is a crucial portion of the NDC80 complex that participates in spindle checkpoint activity and kinetochore-microtubule clustering (4). Recent research has shown that upregulated SPC25 accelerates the proliferation of hepatocellular carcinoma (HCC) and is related to a poor prognosis (5-7). Additionally, previous studies have also reported that SPC25 is an important tumor promoter for cell cycle and malignant progression, and stem cell proliferation in prostate cancer (8). Further, SPC25 has been shown to regulate the stemness of lung cancer cells significantly (9).
Long non-coding ribonucleic acids (lncRNAs) belong to a kind of non-coding RNA that modulates gene transcription or protein translation by multiple distinct mechanisms (10). Alterations in lncRNA expression may exhibit anti-oncogenic or oncogenic activities. The lncRNA cyclin-dependent kinase inhibitor 2B antisense RNA 1 (CDKN2B-AS1) is located at chromosome 9p21, and increases the malignancy of renal cancer (11). Additionally, CDKN2B-AS1 has been shown to induce the proliferation and migration of colorectal cancer (12).
In this study, we found that upregulated SPC25 had an oncogenic function and predicted a poor overall survival (OS) time. Notably, CDKN2B-AS1-activated SPC25 enhanced the proliferation of TNBC cells. Consequently, our research findings may enable the exploration of novel targets for TNBC treatment and prognostic evaluation. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2900/rc).
Methods
Retrieving data
We downloaded the transcriptome profiles and survival information of TNBC from The Cancer Genome Atlas Program (https://portal.gdc.cancer.gov/). GSE45827 dataset was obtained from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) database. We further investigated the pan-cancer expression profiles based on the Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) (13). A Clinical Proteomic Tumor Analysis Consortium (CPTAC, https://cptac-data-portal.georgetown.edu/) was conducted to confirm the protein expression analysis. The Human Protein Atlas (https://www.proteinatlas.org/) was used to verify protein expression levels in diverse tissues and the subcellular localization of the SPC25 protein. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of DEGs and the weighted gene co-expression network analysis (WGCNA)
R software DESeq2 (14) and limma (15) packages were used to identify the differentially expressed genes (DEGs) of the transcriptome data. A WGCNA algorithm (16) was conducted to identify the gene modules that were highly correlated with TNBC. Gene significance (GS) and module membership (MM) were applied to quantify the features and configurations of the modules. The protein–protein interaction network was constructed by the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) with minimum required interaction score >0.4 and PPI enrichment P value <1.0×10−16. Hub genes of TNBC were identified and visualized by the Molecular Complex Detection (MCODE) plugins in Cytoscape. LncSEA as a comprehensive lncRNA sets resource and enrichment analysis platform was performed to analyze functions of lncRNAs.
Functional enrichment analysis
A gene set enrichment analysis (GSEA) was conducted to explore the key module genes and detect some crucial biological functions, such as cell cycle and cellular signaling pathway. GSEA software was used for the GSEA with the following parameters: number of permutations =1,000; Normalized Enrichment Score (NES) >2; nominal (NOM) P value <0.05; and false discovery rate <0.05. The STRING (https://cn.string-db.org/) database was used to identify the proteins co-expressed with SPC25. Subsequently, a Gene Ontology (GO) pathway enrichment analysis was conducted using the clusterProfiler (17) packages in R software.
Single-cell analysis and survival analysis
CancerSEA (http://biocc.hrbmu.edu.cn/CancerSEA/) was applied to reveal the potential function of SPC25 in BC (18). A survival analysis was performed and visualized using the survival and survminer packages in R software.
Cell culture and transfection
Normal human breast cell lines (MCF10A) and TNBC cell lines (i.e., BT549, MDA-MB-231, and MDA-MB-468) were obtained from American Type Culture Collection (ATCC). All the cell lines were authenticated by short tandem repeat deoxyribonucleic acid (DNA) profiling and tested for mycoplasma (GENEWIZ Co., Ltd., Suzhou, China) within 6 months. Lipofectamine 2,000 (Invitrogen, USA) was used for transfection. The GeneCreate Biological Engineering Company (Wuhan, China) synthesized small-interfering RNA specifically targeting CDKN2B-AS1 (si-CDKN2B-AS1) and corresponding non-specific siRNA (si-NC). The siRNA sequences used in our research were: 5'-CCACAUCCCUUGGAGUAAUTT and AUUACUCCAAGGGAUGUGGTT-3'.
RNA extraction and qRT-PCR
We applied the Trizol reagent (Introvigen, USA) to extract total RNA from the cell lines. Subsequently, the RNAs were reverse transcribed into complementary DNA and quantitative real-time polymerase chain reaction (qRT-PCR) assays were conducted. The 2−ΔΔCt method was used to calculate the expression level of CDKN2B-AS1 and SPC25. The primers were purchased from The GeneCreate Biological Engineering Company (Wuhan, China). The primer sequences of the RNAs are shown in Table S1.
Cell proliferation
Cell counting kit-8 (CCK-8) and transwell assays were used to assess cell proliferation viability and migration. First, the MDA-MB-231 cells were transfected with si-CDKN2B-AS1 or si-NC. Second, the CCK-8 solution (Beyotime, China) was supplemented, and the absorbance value at 450 nm was measured at different time points (i.e., 0, 24, 48, and 72 h).
Cell migration experiments
The transfected cells (2×104/well) were seeded into the upper compartment of a 24-well transwell chamber (Corning Costar, USA). The lower chambers contained 10% fetal bovine serum. After incubation for 48 h, the non-migrated cells were carefully removed from the upper chamber with cotton swabs. We randomly selected the average confluence of the migrated cells from 3 visual fields (captured by a 100× microscope) to count the number of cells.
Cell apoptosis experiments
Cells were harvested and washed twice with PBS buffer, 5 µL of Annexin V-FITC and 5 µL of 7-AAD were added to the cells and incubated for 30 min in the dark. Flow cytometer (BD Accuri TM C6) and BD Accuri TM C6 software used to analyze
All of the above experiments was replicated 3 times in laboratory.
Statistical methods
The statistical analysis was conducted using R (v.4.0.3) software. The DESeq2 package was used to analyze the DEGs of the transcriptome data. The WGCNA package was used to construct the co-expression networks. The limma package was used to calculate the DEGs of the microarray data. The gene expression correlation analyses were conducted using Pearson’s coefficients. GSEA software and the clusterProfiler package was used for the pathway enrichment analysis. The survival package was used for the survival analysis. The mean ± standard deviation were calculated, and the statistical difference was examined by the Student’s t-test. A P value <0.05 was considered statistically significant.
Results
WGCNA and GSEA
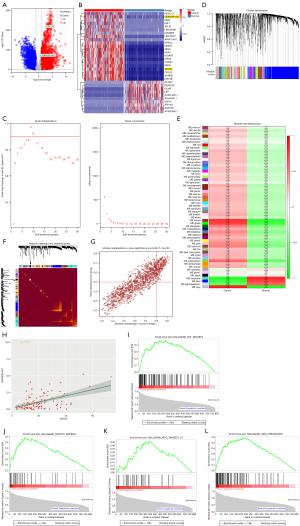
A total of 12,993 DEGs, including lncRNAs, messenger RNAs (mRNAs), and small-nucleolar RNAs, were identified based on the transcriptome data in TCGA (see Figure 1A), and some representative genes were selected to illustrate this different expression patterns (see Figure 1B). The WGCNA were used to identify the gene modules in the para-cancerous tissues and TNBC. To satisfy the scale-free topology, the soft threshold β value was set at 7 (see Figure 1C). In total, 56 gene modules (which were not merged) were constructed by average-linkage hierarchical clustering (see Figure 1D). Additionally, a network heatmap of 600 randomly selected genes revealed a high level of independence across the co-expression clusters (see Figure 1E). The module-trait relationships of different co-expression modules revealed that the brown module showed the highest positive correlation with TNBC (see Figure 1F). In total, 1,155 hub genes were confirmed to have a MM >0.8 and a GS >0.5 in the brown module (available online: https://cdn.amegroups.cn/static/public/atm-22-2900-1.xlsx and Figure 1G). Additionally, in the brown module, CDKN2B-AS1 and SPC25 were more highly expressed in the TNBC tissues than the corresponding normal tissues (see Figure S1A,S1B). The Spearman correlation analysis also revealed that CDKN2B-AS1 and SPC25 were highly and positively correlated (see Figure 1H). STRING database was utilized to enrich the PPI network of DEGs of TNBC. SPC25 as one of hub genes was identified by the MCODE plugin in Cytoscape (see Figure S2A). CDKN2B-AS1 enrichment analysis on LncSEA indicated to be associated with a variety of tumors, including TNBC (see Figure S2B). According to the GSEA, the mRNAs in the brown module were predominantly enriched in the transcription factor E2F and MYC targets, mitotic spindle, and the G2M checkpoint, which are closely associated with cell-cycle regulation (see Figure 1I-1L).
Expression validation, survival analysis, and protein-protein interactions
The expression levels of CDKN2B-AS1 and SPC25 in more than 30 cancer types from TCGA were extracted from GEPIA database. Notably, CDKN2B-AS1 and SPC25 were upregulated in most cancer types (see Figure 2A,2B). Further, the microarray data of GSE45827, comprising 41 basal samples, 14 cell line samples, 30 human epidermal growth factor receptor 2 (Her2+) samples, 29 Luminal A samples, 30 Luminal B samples, and 11 normal samples, were used to validate the upregulation of mRNA SPC25 between the basal samples and normal samples (see Figure 2C). Additionally, the TCGA Cancer Proteome Study of Breast Tissue of The Clinical Proteomic Tumor Analysis Consortium (CPTAC) revealed that the SPC25 protein was highly expressed (see Figure 2D). Subsequently, the SPC25 expression confirmed by the Human Protein Atlas was higher in tumors compared to normal tissue (see Figure 2E,2F). The subcellular location of SPC25 is in the cytoplasm and SPC25 co-locates with microtubule proteins, which play an important role in cell division (see Figure 2G).
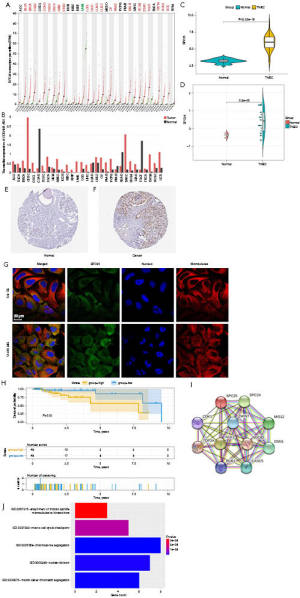
Based on the OS times of TNBC patients in TCGA, a survival analysis of SPC25 was performed to evaluate the difference between the high and low expression groups. Notably, the OS time was lower in patients who had a higher SPC25 expression (see Figure 2H). The protein-protein interaction network of the SPC25 protein was established via the STRING database. The proteins co-expressed with SPC25 were retrieved, and included SPC24, CDK1, ZWINT, MIS12, TOP2A, NUF2, NDC80, DSN1, BUB1, and CASC5 (see Figure 2I). The GO annotation of the co-expressed genes revealed their relevant biological processes (see Figure 2J), which included chromosome segregation, mitotic cell-cycle checkpoint, and nuclear division.
Functions of co-expressed SPC25 and CDKN2B-AS1
Based on the data of the Braune EB (number cells: 369) in CancerSEA, a single-cell analysis was conducted to examine the functions of SPC25, which showed that SPC25 might participate in the regulation of the cell cycle in BC (see Figure 3A,3B). Additionally, the expression of SPC25 was positively correlated with the above-mentioned biological processes (see Figure 3C-3F). The correlation analysis revealed that SPC25 was highly correlated with the cell cycle–related genes (i.e., CDC25A, CCNE1, CCNA2, CCNB1, MKI67, and CDK1) (see Figure 3G).
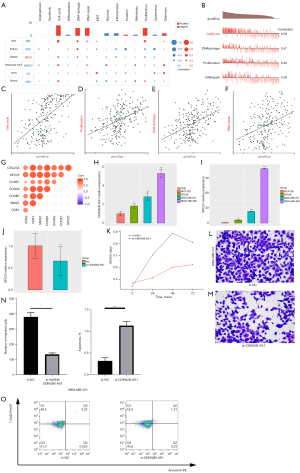
In-vitro studies were used to explore the correlation of SPC25 expression with dysregulated lncRNA CDKN2B-AS1. The validation of the differentially expressed CDKN2B-AS1–SPC25 pair was performed by qRT-PCR. We found that CDKN2B-AS1 and SPC25 were more upregulated in the BT549, MDA-MB-231, and MDA-MB-468 cell lines than the MCF10A cell lines (see Figure 3H,3I). MDA-MB-231 was selected for further interfering assays. As expected, the knockdown of CDKN2B-AS1 inhibited SPC25 expression (see Figure 3J). Further, the cell number monitoring assays revealed that the cell proliferation varied (see Figure 3K) and the invasion ability was restrained (see Figure 3L-3N) after the knockdown of CDKN2B-AS1. In addition, flow cytometry cell apoptosis analysis showed that knockdown of CDKN2B-AS1 significantly increased the percentage of apoptotic cells (see Figure 3O).
Discussion
In our study, a WGCNA was conducted to find the co-expression gene modules, and we identified the key gene modules associated with tumorigenesis in the network. The WGCNA package uses bioinformatics approaches to describe clusters of highly correlated genes (19). We discovered that CDKN2B-AS1 and mRNA SPC25 in the brown module were significantly overexpressed in TNBC. We further characterized the synergistic effect of CDKN2B-AS1 and SPC25 and found that they accelerated TNBC. To examine the effects of the upregulation of CDKN2B-AS1 on TNBC proliferation and migration, CCK-8 and transwell assays were conducted, and revealed that the overexpression of SPC25 was positively correlated with the poor OS of TNBC patients. Further, our findings revealed that CDKN2B-AS1 may promote TNBC proliferation by upregulating SPC25 and accelerating the cell cycle.
Recently, studies have revealed that lncRNAs are aberrantly expressed in breast cancer and closely associated with the development and progression of breast cancer (20). LncRNAs can regulate mRNA expression through multiple mechanisms: epigenetic regulation, transcriptional regulation, post-transcriptional regulation, acting as ceRNA and other molecules mechanisms (21). However, the specific mechanism is still unclear and it is a challenge to use lncRNAs as biomarkers for diagnosis or therapeutic targets. We found that CDKN2B-AS1 and mRNA SPC25 were significantly overexpressed in TNBC. LncRNA CDKN2B-AS1 participates in a variety of regulatory roles, including the cell cycle of tumors (11,22-24). The aberrant expression of CDKN2B-AS1 has been reported in multiple cancers, and it might accelerate proliferation and inhibit the apoptosis of tumor cells (25). Studies have revealed that the mRNA level of CDKN2B‑AS1 is increased and exerts carcinogenic effects in various tumors (11,26,27). Research has shown that CDKN2B-AS1 is overexpressed in gliomas and enhances cell proliferation, invasion, and migration, and inhibits the apoptosis of glioma cells (28). CDKN2B-AS1 has also been shown to be significantly overexpressed in renal carcinoma cells, while the downregulation of CDKN2B-AS1 has been shown to suppress the tumor growth involved in proliferation, migration, and invasion (29).
As a part of the Ndc80 complex, the SPC25 homolog plays an important role in the mitosis of cell-cycle processes, including spindle formation and chromosome alignment (30-32). SPC25 and CDKN2B-AS1 expression levels are upregulated in most of the cancers identified by the GEPIA database, and the GEO analysis confirmed that the expression of SPC25 was significantly more elevated in TNBC tissues than normal tissues. SPC25 has been shown to be upregulated and involved in cell proliferation in lung cancer (33). We found that the expression of SPC25 was downregulated by siRNA-mediated CDKN2B‑AS1 knockdown. Additionally, the Spearman correlation analysis revealed that the expression levels of SPC25 and CDKN2B-AS1 were highly and positively correlated. The CCK-8 and transwell assays indicated that proliferation and migration were strongly inhibited in the si-CDKN2B‑AS1 cell line. We confirmed the synergistic effect of CDKN2B-AS1 and that SPC25 enhances the proliferation and migration capacity of TNBC. Similarly, previous studies have demonstrated that the downregulation of SPC25 significantly reduces the proliferation of prostate cancer (34). Additionally, SPC25 has been shown to interact with checkpoint-related proteins and increase proliferation in lung cancer (33). SPC25 facilitates tumor growth via the tumor protein P53 pathway and may be a novel tumor-promoting factor and a therapeutic target in HCC (7). The downregulation of SPC25 also leads to aberrant spindle organization with misaligned chromosomes (31). Thus, we hypothesize that SPC25 overexpression is involved in genetic alterations in the cell cycle caused by incorrect chromosome segregation, in which motility plays a crucial role, and ultimately leads to tumorigenesis. We also observed that SPC25 was colocalized with microtubule proteins. Previous research has shown that tubulin plays a vital role in cell proliferation (35). Thus, we hypothesize that SPC25 binds to microtubule proteins and then accelerates TNBC tumorigenesis and progression.
Next, the survival analysis confirmed that the SPC25 level was an independent prognostic factor of TNBC. High levels of SPC25 were related to poor OS and recurrence-free survival in lung adenocarcinoma patients (9). Research has indicated that increased SPC25 expression levels are correlated with poor OS in BC patients (36). The high expression of SPC25 has also been shown to be correlated with short survival time and to enhance the proliferation of HCC cells (5). The patient with higher SPC25 expression had higher recurrence rates, which was associated with shorter OS in BC patients (37). Our study was the first to reveal that SPC25 could have potential clinical value in TNBC.
In this study, the GO annotation of the co-expressed genes identified the relevant biological processes, including chromosome segregation, mitotic cell-cycle checkpoint, and nuclear division. We performed a GSEA to evaluate the underlying function of the brown module. WGCNA and pathway analyses have shown that SPC25 is a potential marker of HCC and is associated with the mitotic cell cycle (38,39). Genes in the brown module were primarily related to cell-cycle regulation, which is a pervasive and complex sequence of events (40). Aberrant cell cycles, which allow cells to divide indefinitely, are hallmarks of cancer. Thus, therapies targeting cell cycles have long been regarded as promising anti-cancer strategies. Genes in the brown module may potentially promote tumor development via the transcription of E2F and MYC to accelerate cell cycles. In the future, the synergistic effect of CDKN2B-AS1 and SPC25 might be important for improving the optimal development and application of cell-cycle targeting drugs in clinical practice.
The single-cell analysis demonstrated that SPC25 may contribute to the progression of BC by accelerating the cell cycle. The downregulation of SPC25 expression is associated with cell-cycle at M-phase arrest (32). The correlation analysis revealed that SPC25 may increase the cell cycle through co-expression with CDC25A, CCNE1, CCNA2, CCNB1, MKI67, and CDK1. Further experiments need be conducted to verify the interaction of SPC25 with these cell cycle-related genes.
In summary, our research shows that the synergistic effect of CDKN2B-AS1 and SPC25 on the occurrence and development of TNBC. CDKN2B-AS1 may play a significant role in the diagnosis, prognosis, and specific therapeutic target in TNBC. Our results provided new insights into the potential biomarkers and mechanisms of TNBC oncogenesis and also suggest a potential application of treatment in the future.
This study had a number of limitations. First, while CDKN2B‑AS1 and SPC25 are potential markers, large-scale validations studies urgently need to be conducted to apply these findings in a clinical setting. Second, the WGCNA method has limitations, as its selection criteria and thresholds may affect the ultimate findings.
Conclusions
In brief, we used the WGCNA method to identify the synergistic effect of lncRNAs and mRNAs on TNBC progression. We also verified the functional roles and molecular process of CDKN2B‑AS1 as a tumor-promoting factor that upregulates SPC25 expression in tumor tissues. Collectively, we identified a number of TNBC co-expression modules and hub genes. Our findings provide a theoretical foundation for future research. The candidate factors may be applied as diagnostic and prognostic biomarkers or clinical treatment targets in the future.
Acknowledgments
The authors would like to thank TCGA and the GEO databases and all the contributors to the databases who have provided open-source data for researchers.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-2900/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-2900/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2900/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Szymiczek A, Lone A, Akbari MR. Molecular intrinsic versus clinical subtyping in breast cancer: A comprehensive review. Clin Genet 2021;99:613-37. [Crossref] [PubMed]
- de Paula BHR, Kumar S, Morosini FM, et al. Real-world assessment of the effect of impact of tumor size on pathological complete response rates in triple negative breast cancer after neoadjuvant chemotherapy. Chin Clin Oncol 2020;9:78. [Crossref] [PubMed]
- Janke C, Ortiz J, Lechner J, et al. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J 2001;20:777-91. [Crossref] [PubMed]
- Zhang B, Zhou Q, Xie Q, et al. SPC25 overexpression promotes tumor proliferation and is prognostic of poor survival in hepatocellular carcinoma. Aging (Albany NY) 2020;13:2803-21. [Crossref] [PubMed]
- Yang X, Sun H, Song Y, et al. Diagnostic and prognostic values of upregulated SPC25 in patients with hepatocellular carcinoma. PeerJ 2020;8:e9535. [Crossref] [PubMed]
- Chen F, Zhang K, Huang Y, et al. SPC25 may promote proliferation and metastasis of hepatocellular carcinoma via p53. FEBS Open Bio 2020;10:1261-75. [Crossref] [PubMed]
- Cui F, Tang H, Tan J, et al. Spindle pole body component 25 regulates stemness of prostate cancer cells. Aging (Albany NY) 2018;10:3273-82. [Crossref] [PubMed]
- Chen J, Chen H, Yang H, et al. SPC25 upregulation increases cancer stem cell properties in non-small cell lung adenocarcinoma cells and independently predicts poor survival. Biomed Pharmacother 2018;100:233-9. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- Xie X, Lin J, Fan X, et al. LncRNA CDKN2B-AS1 stabilized by IGF2BP3 drives the malignancy of renal clear cell carcinoma through epigenetically activating NUF2 transcription. Cell Death Dis 2021;12:201. [Crossref] [PubMed]
- Zheng Y, Zeng J, Xia H, et al. Upregulated lncRNA Cyclin-dependent kinase inhibitor 2B antisense RNA 1 induces the proliferation and migration of colorectal cancer by miR-378b/CAPRIN2 axis. Bioengineered 2021;12:5476-90. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Yuan H, Yan M, Zhang G, et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res 2019;47:D900-D8. [Crossref] [PubMed]
- Song ZY, Chao F, Zhuo Z, et al. Identification of hub genes in prostate cancer using robust rank aggregation and weighted gene co-expression network analysis. Aging (Albany NY) 2019;11:4736-56. [Crossref] [PubMed]
- Jin H, Du W, Huang W, et al. lncRNA and breast cancer: Progress from identifying mechanisms to challenges and opportunities of clinical treatment. Mol Ther Nucleic Acids 2021;25:613-37. [Crossref] [PubMed]
- Arencibia A, Lanas F, Salazar LA. Long Non-Coding RNAs Might Regulate Phenotypic Switch of Vascular Smooth Muscle Cells Acting as ceRNA: Implications for In-Stent Restenosis. Int J Mol Sci 2022;23:3074. [Crossref] [PubMed]
- Alexander JL, Wilson ID, Teare J, et al. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol 2017;14:356-65. [Crossref] [PubMed]
- Ali MW, Patro CPK, Devall M, et al. A Functional Variant on 9p21.3 Related to Glioma Risk Affects Enhancer Activity and Modulates Expression of CDKN2B-AS1. Hum Mutat 2021;42:1208-14. [Crossref] [PubMed]
- Pan J, Lin M, Xu Z, et al. CDKN2B antisense RNA 1 suppresses tumor growth in human colorectal cancer by targeting MAPK inactivator dual-specificity phosphatase 1. Carcinogenesis 2021;42:1399-409. [Crossref] [PubMed]
- Song C, Qi Y, Zhang J, et al. CDKN2B-AS1: An Indispensable Long Non-coding RNA in Multiple Diseases. Curr Pharm Des 2020;26:5335-46. [Crossref] [PubMed]
- Qin S, Ning M, Liu Q, et al. Knockdown of long non-coding RNA CDKN2B-AS1 suppresses the progression of breast cancer by miR-122-5p/STK39 axis. Bioengineered 2021;12:5125-37. [Crossref] [PubMed]
- Yang D, Ma J, Ma XX. CDKN2B-AS1 Promotes Malignancy as a Novel Prognosis-Related Molecular Marker in the Endometrial Cancer Immune Microenvironment. Front Cell Dev Biol 2021;9:721676. [Crossref] [PubMed]
- Lu J, Chen Y, Wen L, et al. LncRNA CDKN2B-AS1 contributes to glioma development by regulating the miR-199a-5p/DDR1 axis. J Gene Med 2022;24:e3389. [Crossref] [PubMed]
- Dasgupta P, Kulkarni P, Majid S, et al. LncRNA CDKN2B-AS1/miR-141/cyclin D network regulates tumor progression and metastasis of renal cell carcinoma. Cell Death Dis 2020;11:660. [Crossref] [PubMed]
- Xiong B. Spc25: How the kinetochore protein plays during oocyte meiosis. Cell Cycle 2011;10:1031-0. [Crossref] [PubMed]
- Sun SC, Lee SE, Xu YN, et al. Perturbation of Spc25 expression affects meiotic spindle organization, chromosome alignment and spindle assembly checkpoint in mouse oocytes. Cell Cycle 2010;9:4552-9. [Crossref] [PubMed]
- McCleland ML, Kallio MJ, Barrett-Wilt GA, et al. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr Biol 2004;14:131-7. [Crossref] [PubMed]
- Jeong J, Keum S, Kim D, et al. Spindle pole body component 25 homolog expressed by ECM stiffening is required for lung cancer cell proliferation. Biochem Biophys Res Commun 2018;500:937-43. [Crossref] [PubMed]
- Cui F, Hu J, Fan Y, et al. Knockdown of spindle pole body component 25 homolog inhibits cell proliferation and cycle progression in prostate cancer. Oncol Lett 2018;15:5712-20. [Crossref] [PubMed]
- Tangutur AD, Kumar D, Krishna KV, et al. Microtubule Targeting Agents as Cancer Chemotherapeutics: An Overview of Molecular Hybrids as Stabilizing and Destabilizing Agents. Curr Top Med Chem 2017;17:2523-37. [Crossref] [PubMed]
- Pathania R, Ramachandran S, Mariappan G, et al. Combined Inhibition of DNMT and HDAC Blocks the Tumorigenicity of Cancer Stem-like Cells and Attenuates Mammary Tumor Growth. Cancer Res 2016;76:3224-35. [Crossref] [PubMed]
- Wang Q, Zhu Y, Li Z, et al. Up-regulation of SPC25 promotes breast cancer. Aging (Albany NY) 2019;11:5689-704. [Crossref] [PubMed]
- Gu Y, Li J, Guo D, et al. Identification of 13 Key Genes Correlated With Progression and Prognosis in Hepatocellular Carcinoma by Weighted Gene Co-expression Network Analysis. Front Genet 2020;11:153. [Crossref] [PubMed]
- Wan Z, Zhang X, Luo Y, et al. Identification of Hepatocellular Carcinoma-Related Potential Genes and Pathways Through Bioinformatic-Based Analyses. Genet Test Mol Biomarkers 2019;23:766-77. [Crossref] [PubMed]
- Poon RY. Cell Cycle Control: A System of Interlinking Oscillators. Methods Mol Biol 2016;1342:3-19. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


