
CRISPR/Cas9 ribonucleoprotein (RNP) complex enables higher viability of transfected cells in genome editing of acute myeloid cells
Introduction
Acute myeloid leukemia (AML) is a malignant proliferation of abnormal myeloid cells and has an intermediate risk of recurrence (1). Although remarkable advances have been made in hematologic tumor therapy, the majority of AML patients still suffer from relapse after extensive treatment. AML cells are nonadherent cells and the molecular biological approaches for studying AML are very different from those for solid tumors. AML cells are difficult to transfect or transduce, which is the main limitation of AML research and therapy.
The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system has become an increasingly vital tool for modifying gene expression in a variety of cell types, showing unprecedented potential for tumor therapy (2). Many studies have applied CRISPR/Cas9 to therapeutically relevant ex vivo and in vivo experiments (3). However, the application of CRISPR/Cas9 is restricted because of the large size of the Cas9 protein (160 kDa). Efficient and safe delivery of CRISPR/Cas9 into AML cells is urgently needed to explore new therapeutic targets. In recent years, CRISPR/Cas9 ribonucleoprotein (RNP) complex has been directly delivered into tumor cells via electroporation, leading to decreased off-target effects due to the short lifetime of the RNP complex (4-9).
Electroporation is an electro-physical method for gene and protein transfer based on square-wave pulse technology and results in high expression of delivery targets and substantial viability of cells (10). Electroporation is already successfully applied to the manufacture of antitumor lymphocytes (11) and delivery of exogenous drugs and nuclear acids (12). CRISPR/Cas9 with electroporation shows great potential for genome editing and tumor therapy. However, the fundamental mechanism of electroporation-mediated DNA/RNA transfer has not yet been fully elucidated. The electric parameters (pulse amplitude, volts, etc.) need to be optimized for various conditions to reduce cell death and increase transfection efficiency (13).
Lentivirus or nanoparticles has been used to deliver CRISPR/Cas9 system into cells in many research studies, but the cell viability or transfection efficiency was always challenging. In this study, electroporation was used to deliver the CRISPR/Cas9 system, including CRISPR/Cas9 all-in-one plasmid and the RNP complex. Compared to CRISPR/Cas9 all-in-one plasmid, the RNP complex enabled higher viability of transfected cells. A novel fluorescent labeling of RNPs was applied to select genome-edited AML cells, which substantially improved the genome editing efficiency. Furthermore, gene editing of CD34+ cells was also carried out in our study. This electroporating approach to deliver CRISPR/Cas9 RNP is simple and easy to operate, which provides a potential option for cancer research and tumor therapeutics. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3279/rc).
Methods
Cell culture
Human leukemia cell lines MV4-11 and Kasumi-1 were purchased from the American Type Culture Collection (ATCC; Rockville, MD, USA). MV4-11 and Kasumi-1 cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium supplemented with 10% and 20% fetal bovine serum, respectively. Cell density was kept between 1×105 and 1×106 cells/mL. Cells were cultured at 37 ℃ in a humid incubator supplied with 5% CO2.
Primary CD34+ cells were isolated from cord blood with donors’ consent using the commercial isolation kit (STEMCELL Technologies, EasySepTM Human CD34 Positive Selection Kit II, Canada). CD34+ cells were cultured in StemSpan SFEM II (STEMCELL Technologies, Vancouver, BC, Canada) supplemented with FMS-like tyrosine kinase 3 ligand (FLT3L; 100 ng/mL), thrombopoietin (100 ng/mL), and stem cell factor (100 ng/mL; PeproTech, East Windsor, NJ, USA). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all the patients. The study was approved by institutional ethics board of Shanghai Tenth People’s Hospital of Tongji University (22KN250).
Cloning
Guide DNAs (gDNAs) targeting enhanced green fluorescent protein (EGFP; sequence: GGCCACAAGTTCAGCGTGTC) was synthesized by TSINGKE Biotechnology Co., Ltd. (Beijing, China). The double strands DNAs were denatured at 95 ℃ for 5 minutes, and then step-wise annealing was carried out. Simultaneously, the pL-CRISPR.EFS.GFP (57818; Addgene, Waterton, MA, USA) vector was digested via FastDigest Esp3I (Thermo Fisher Scientific, Waltham, MA, USA) at 37 ℃ for 15 minutes and purified on a 1% agarose gel (Biozyme). T4 DNA ligase (Thermo Fisher Scientific) was used to ligate the annealed double-stranded gDNA and digested vector (5:1 molar ratio). The ligated product was transformed into 50 µL of Stbl3 competent cells. Correct clones were confirmed by Sanger sequencing.
Guide RNA (gRNA) in vitro transcription
Plasmid pL-CRISPR.EFS.GFP was used as a DNA template for gRNA in vitro transcription. The forward primer was composed of forward primers containing a T7 promoter sequence (TTAATACGACTCACTATAG), the specific targeting sequence, and the sequence corresponding to the 5' terminal of the gRNA scaffold (GTTTTAGAGCTAGAAATAGC). The reverse primer sequence was: AGCACCGACTCGGTGCCACT. The EGFP targeting sequence was: GGAGCGCACCATCTTCTTCA (14). Polymerase chain reaction (PCR) products were purified with Gel DNA Recovery Kit (Thermo Fisher Scientific). HiScribe T7 High Yield RNA Synthesis Kit [New England Biolabs (NEB) GmbH, Frankfurt, Germany] was used to perform the in vitro transcription according to the manufacturer instructions. The transcribed gRNA products were purified through DNA electrophoresis and were recovered with RNA Clean & Concentrator Kit (Thermo Fisher Scientific).
Electroporation
Purified in vitro transcribed gRNA (1 µg) was incubated with 1 µg Cas9 protein (PNA Bio, Newbury Park, CA, USA) at room temperature for 15 minutes before electroporation. Neon transfection system (Thermo Fisher Scientific) was used and the various electroporation parameters were tested, the electroporating voltage which have been tried in this study changed from 900 to 1,600 v. Electroporation in Buffer R (Invitrogen, Carlsbad, CA, USA) was performed for 3 independent experiments, and 150,000–250,000 cells were used in each replicate. After 96 hours, the genome editing efficiency was validated.
Cell viability assays
The MV4-11 cells, Kasumi-1 cells, and CD34+ cells were electroporated with CRISPR/Cas9-GFP all-in-one plasmid and the RNP complex, respectively, with different electroporation parameters according to the aforementioned method. After 96 hours, cells were stained with trypan blue and counted under microscopy. All experiments were repeated at least 3 times.
Electrophoretic mobility shift assay (EMSA)
After incubation of purified gRNA (1 µg) and Cas9 protein (1 µg) at room temperature for 15 minutes, gRNA/Cas9 RNP was loaded on 1.5% agarose gel to check the combining efficiency and electrophoresis was performed in 1× tris-acetate-EDTA (TAE) buffer with the Bio-Rad Powersupplies (130 v). Agarose gel imager was used to document the results.
gRNA 3' end labeling
pCp-Cy5 (30 µM; Sangon Biotechnology, Shanghai, China) was ligated to 1 µg gRNA with T4 RNA ligase (NEB) supplied with 10 mM adenosine triphosphate (ATP) at 16 ℃ overnight. The labeled RNA was purified with RNA Clean & Concentrator-5 (cat. No. R1013; Zymo Research, Irvine, CA, USA). For each experiment, fresh fluorescent labeled gRNA should be prepared in this study.
Flow cytometry analysis
Transfected Cy5-positive cells were sorted by fluorescence-activated cell sorting (FACS; BD FACSAria II; BD Biosciences, Franklin Lakes, NJ, USA) and 1× PBS with 2% FBS were used to collect the sorted cells. The genome editing efficiency was then tested through GFP expression under flow cytometry (BD LSR Flow Cytometer, BD Sciences).
Statistical analyses
The data were analyzed with SPSS version 22 (IBM Corp., Armonk, NY, USA). Student’s t-test was used to confirm the statistical significance, and P<0.05 was considered statistically significant.
Results
CRISPR/Cas9 RNP reveals higher viability
To test the efficiency of electroporation on cell viability, we performed electroporation of leukemia cells with CRISPR/Cas9 all-in-one plasmid and gRNA/Cas9 RNPs. When plasmids were transfected into MV4-11 cells, cell viability was only around 5%, while it was up to 10 times higher (45–61%) when RNPs were delivered (Figure 1A). For Kasumi-1 cells, when plasmids were electroporated, a relatively higher number of living cells (compared to MV4-11 cells) were observed. Notably, when gRNA/Cas9 RNPs were used, viability was also significantly increased (Figure 1B). These results indicated that RNPs were much less toxic to cells, and electroporation with gRNA/Cas9 RNPs was a more viable and acceptable approach.
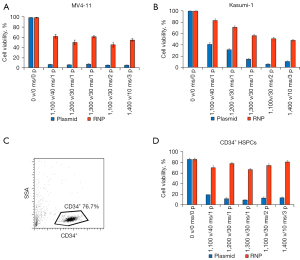
It is well known that hematopoietic stem and progenitor cells (HSPCs) are difficult to transfect with conventional viral and nonviral methods. Herein, we isolated CD34+ HSPCs from cord blood (Figure 1C) and electroporated these cells with CRISPR/Cas9 all-in-one plasmid and gRNA/Cas9 RNPs, respectively. Consistent with the aforementioned results, the cells electroporated with gRNA/Cas9 RNPs showed much higher viability than those with CRISPR/Cas9 all-in-one plasmid (Figure 1D). These results collectively identified the higher viability of nonadherent cells after electroporating gRNA/Cas9 RNPs and the potential high delivery efficiency of gRNA/Cas9 RNPs.
Fluorescent labeling CRISPR/Cas9 RNPs enable effective genome editing
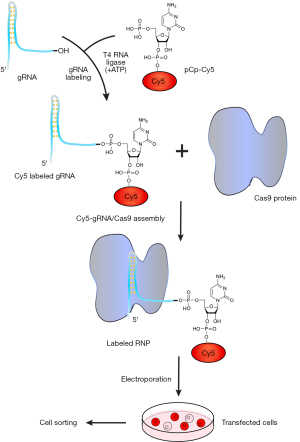
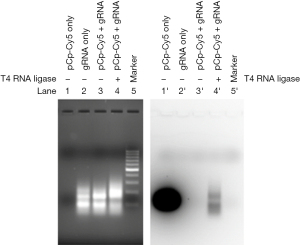
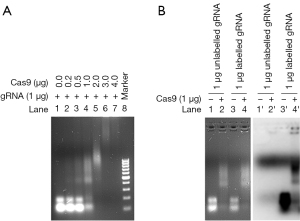
As selecting cells transfected with gRNA/Cas9 RNPs is difficult, we designed an easy and effective technique for isolating cells with introduced RNPs. We first labeled in vitro transcribed gRNA with pCp-Cy5 via T4 ligase before incubating with Cas9 protein, as shown in the gRNA/Cas9 RNP labeling schematic (Figure 2). Cy5 labeling of gRNA was verified using Imager 600 with red fluorescence (Figure 3). Neither gRNA alone (lane 2’) nor pCp-Cy5 plus gRNA without T4 RNA ligase (lane 3’) displayed a detectable band, while pCp-Cy5 plus gRNA with T4 RNA ligase (lane 4’) showed a clear band, indicating the labeling was successful and specific. Further, the in vitro transcribed gRNA could effectively bind Cas9 protein in a proportion-dependent manner (Figure 4A). In order to test the possible effect of Cy5 labeling on the binding of gRNA to Cas9 protein, EMSA was performed. The results indicated that Cy5 labeling had no obvious effect on the binding of gRNA to Cas9 protein (Figure 4B). The above results confirmed the successful production of labeled gRNA/Cas9 RNPs in vitro.
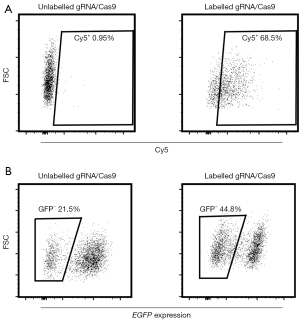
To further explore the efficiency of CRISPR/Cas9 RNPs on genome editing, Cy5-labeled gRNA/Cas9 RNPs were electroporated into the MV4-11 cells with EGFP under the electric parameters of 1,600 v, 10 ms, and 3 pulses. The expression of targeted gene (GFP) was significantly downregulated in the Cy5-sorted cells (44.8%) compared to the unsorted cells (21.5%) (Figure 5). In sum, our results identified the substantial benefits of fluorescent labeling CRISPR/Cas9 RNPs on genome editing of nonadherent cells.
Discussion
Recently, nonviral delivery systems for DNA-molecule transport have become increasingly important due to the risk of insertional mutagenesis and probable immunogenicity of viral systems (15). Electroporation has been widely used in gene delivery for vaccine production, transgene expression, and therapy for various cancers (16). As nonadherent cells, AML cells are much more difficult to transfect compared to adherent cells, even with a viral delivery system. The applied range of electroporation is broad and flexible, depending on the transported molecule (mRNA, DNA, protein, etc.) and the various target cell types (adherent and nonadherent cells) (17). Herein, electroporation, which has been used for the past 30 years, was chosen as the delivery method for CRISPR/Cas9 RNPs. The transport efficiency of CRISPR/Cas9 RNPs into AML cells with electroporation is high and cell death is low, confirming the feasibility of electroporation in AML cells.
Traditionally, gene editing has been used to investigate the function of targeted genes and develop novel ways for treating diseases, especially malignancies. A number of gene editing approaches have been used over the past decades, including zinc-finger nucleases, transcription activator-like effector nucleases, and so on (18). Recently, gene editing using CRISPR/Cas9 has quickly risen in popularity due to its simplicity and speed. CRISPR/Cas9, working through RNA-guided Cas9 nucleases, is an efficient tool for gene editing. However, viral-delivered CRISPR/Cas9 plasmids normally result in prolonged expression of the gene editing system, which often increases the frequency of off-target effects and promotes oncogenesis in transduced cells (5). As a result, CRISPR/Cas9 RNPs, with their short lifespan, have been synthesized, and various approaches for delivering CRISPR/Cas9 RNPs have been developed to overcome the drawbacks of viral delivery (19-21). Consistent with previous reports, our results indicated that electroporated CRISPR/Cas9 plasmids into AML cells induced high cytotoxicity. Conversely, CRISPR/Cas9 RNPs delivered with electroporation into AML cells led to more cell viability. Furthermore, Cy5 labeling of gRNA/Cas9 RNPs was used to select positive cells, which could significantly enhance gene editing efficiency. Previous studies have used nanoparticles to deliver CRISPR/Cas9 RNPs, and approximate 30% indel efficiency of target gene have been identified (7), which is significantly lower than that via electroporation (44.8%). Our results indicated that the use of Cy5-labeled CRISPR/Cas9 RNPs could be beneficial for gene editing in nonadherent cells. Of note, the specific mutations generated by CRISPR/Cas9 was mediated by both NHEJ- and HDR-based genome modifications (22). The off-target mutations is normally serious concern about CRISPR/Cas9, which mainly depends on the specificity of targeting regions. To minimize the off-target of gene editing, the tools (http://www.xn--genome-engineering-rl9j.org/crispr/?page_id=41) has been used to design the sgRNA and normally more than one sgRNAs will be recommended in research study. So far, the application of CRISPR/Cas9 RNPs for clinical gene editing is still challenging.
Over the last few years, molecularly targeted inhibitors and chemotherapy drugs have been used to treat malignancies, improving the quality of life for patients (3). However, drug resistance often leads to recurrence. Gene editing, especially CRISPR/Cas9-mediated gene editing, has paved the way for new approaches to cancer therapy. Nanoparticle delivery of CRIPR/Cas9 has been used to rescue fragile X syndrome in mouse model (8), and lentivirus-delivered CRIPR/Cas9 has been confirmed the positive therapeutic role on malignancy (23-26). However, due to the off-target effects of CRISPR/Cas9 and the delivery vehicle, its application has been limited. As CRISPR/Cas9 RNPs degrade in cells, excessive cleavage of untargeted genes is avoided, and thus electroporated CRISPR/Cas9 RNPs may provide a new option for cancer therapy.
Acknowledgments
Funding: This work was supported by the Shanghai Sailing Program (No. 21YF1435300), the Project of Key Laboratory of Endemic and Ethnic Diseases, Ministry of Education, Guizhou Medical University (No. FZSW-2021-005), the Shanghai Municipal Health Commission (No. 20194Y001).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3279/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3279/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3279/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all the patients. The study was approved by institutional ethics board of Shanghai Tenth People’s Hospital of Tongji University (22KN250).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferraro F, Miller CA, Christensen KA, et al. Immunosuppression and outcomes in adult patients with de novo acute myeloid leukemia with normal karyotypes. Proc Natl Acad Sci U S A 2021;118:e2116427118. [Crossref] [PubMed]
- Wright AV, Nuñez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature's Toolbox for Genome Engineering. Cell 2016;164:29-44. [Crossref] [PubMed]
- Rosenblum D, Gutkin A, Kedmi R, et al. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci Adv 2020;6:eabc9450. [Crossref] [PubMed]
- Gee P, Lung MSY, Okuzaki Y, et al. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat Commun 2020;11:1334. [Crossref] [PubMed]
- Banskota S, Raguram A, Suh S, et al. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell 2022;185:250-265.e16. [Crossref] [PubMed]
- Newby GA, Yen JS, Woodard KJ, et al. Base editing of haematopoietic stem cells rescues sickle cell disease in mice. Nature 2021;595:295-302. [Crossref] [PubMed]
- Mout R, Ray M, Yesilbag Tonga G, et al. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano 2017;11:2452-8. [Crossref] [PubMed]
- Lee B, Lee K, Panda S, et al. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat Biomed Eng 2018;2:497-507. [Crossref] [PubMed]
- Vakulskas CA, Dever DP, Rettig GR, et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med 2018;24:1216-24. [Crossref] [PubMed]
- Chicaybam L, Barcelos C, Peixoto B, et al. An Efficient Electroporation Protocol for the Genetic Modification of Mammalian Cells. Front Bioeng Biotechnol 2016;4:99. [PubMed]
- Ramanayake S, Bilmon I, Bishop D, et al. Low-cost generation of Good Manufacturing Practice-grade CD19-specific chimeric antigen receptor-expressing T cells using piggyBac gene transfer and patient-derived materials. Cytotherapy 2015;17:1251-67. [Crossref] [PubMed]
- Sokołowska E, Błachnio-Zabielska AU. A Critical Review of Electroporation as A Plasmid Delivery System in Mouse Skeletal Muscle. Int J Mol Sci 2019;20:2776. [Crossref] [PubMed]
- Sherba JJ, Hogquist S, Lin H, et al. The effects of electroporation buffer composition on cell viability and electro-transfection efficiency. Sci Rep 2020;10:3053. [Crossref] [PubMed]
- Shalem O, Sanjana NE, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014;343:84-7. [Crossref] [PubMed]
-
Karagöz U Kotmakçı M Akbaba H Preparation and characterization of non-viral gene delivery systems with pEGFP-C1 Plasmid DNA. Braz J Pharm Sci 2018 . doi: . - Young JL, Dean DA. Electroporation-mediated gene delivery. Adv Genet 2015;89:49-88. [Crossref] [PubMed]
- Cao Y, Ma E, Cestellos-Blanco S, et al. Nontoxic nanopore electroporation for effective intracellular delivery of biological macromolecules. Proc Natl Acad Sci U S A 2019;116:7899-904. [Crossref] [PubMed]
- Martinez-Lage M, Puig-Serra P, Menendez P, et al. CRISPR/Cas9 for Cancer Therapy: Hopes and Challenges. Biomedicines 2018;6:105. [Crossref] [PubMed]
- Zhang S, Shen J, Li D, et al. Strategies in the delivery of Cas9 ribonucleoprotein for CRISPR/Cas9 genome editing. Theranostics 2021;11:614-48. [Crossref] [PubMed]
- Lee K, Conboy M, Park HM, et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat Biomed Eng 2017;1:889-901. [Crossref] [PubMed]
- Wang P, Zhang L, Xie Y, et al. Genome Editing for Cancer Therapy: Delivery of Cas9 Protein/sgRNA Plasmid via a Gold Nanocluster/Lipid Core-Shell Nanocarrier. Adv Sci (Weinh) 2017;4:1700175. [Crossref] [PubMed]
- Ran FA, Hsu PD, Wright J, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8:2281-308. [Crossref] [PubMed]
- Cui C, Liu Y, Gerloff D, et al. NOP10 predicts lung cancer prognosis and its associated small nucleolar RNAs drive proliferation and migration. Oncogene 2021;40:909-21. [Crossref] [PubMed]
- Kim W, Lee S, Kim HS, et al. Targeting mutant KRAS with CRISPR-Cas9 controls tumor growth. Genome Res 2018;28:374-82. [Crossref] [PubMed]
- Moreno AM, Fu X, Zhu J, et al. In Situ Gene Therapy via AAV-CRISPR-Cas9-Mediated Targeted Gene Regulation. Mol Ther 2018;26:1818-27. [Crossref] [PubMed]
- Rafii S, Tashkandi E, Bukhari N, et al. Current Status of CRISPR/Cas9 Application in Clinical Cancer Research: Opportunities and Challenges. Cancers (Basel) 2022;14:947. [Crossref] [PubMed]
(English Language Editor: A. Muijlwijk)


