
Proscillaridin A induces mitochondrial damage and autophagy in pancreatic cancer and reduces the stability of SMAD4 in Panc-1 cells
Introduction
Pancreatic cancer (PC) is one of the most aggressive and lethal cancers (1). Previous studies have shown that aging, environmental and behavioral changes are more closely related to the dramatic increase in the incidence of PC worldwide than genetic factors (2-5). A position paper published by the European Federation of Gastroenterology described PC as a medical emergency (6).
In the past few years, first- and second-line palliative systemic treatments and adjuvant therapies have made considerable progress in prolonging survival of PC patients. The combination therapy composed of folic acid, 5-fluorouracil, irinotecan and oxaliplatin increased the overall survival of patients with PC by 4.3 months (7,8). In addition, nab-paclitaxel combined with gemcitabine can improve the median progression-free survival rate and objective remission rate (9). However, the prognosis of PC is still very poor, and the 5-year survival rate is lower than 5% (10). There is an urgent need for new drugs and strategies against PC.
PC is characterized by complex genetic alterations. Several signaling pathways have been reported to play an important role in the pathogenesis of PC by regulating the proliferation and apoptosis of cancer cells (11). Mutations in at least 32 genes belonging to 10 cellular signaling pathways are associated with PC (12). Four genes, KRAS, CDKN2A, TP53, and SMAD4, are the most commonly mutated genes in PC (13-16), which may help to guide personalized cancer therapy. A previous study showed that the decrease in Ras activity could inhibit the proliferation of KRAS wild-type PC cells and increase the sensitivity to cetuximab (17).
Proscillaridin A (Pro A), one of the cardiac glycosides (CGs), has long been used to treat congestive heart failure, especially in the case of bradycardia, where its negative chronotropic effect is considered to be less than that of digitalis (18). In cardiomyocytes, Pro A binds and inhibits Na+/K+-ATPase, resulting in enhanced myocardial contraction (19). Pro A has shown anti-tumor effects on various cancers, including breast cancer (20), prostate cancer (21), hepatocellular carcinoma (22), leukemia (23) and non-small cell lung cancer (24). Pro A has also been demonstrated to inhibit PC cells at micro-molar concentrations using 3D spheroid cultures (25). The mechanism of Pro A inhibiting PC cells is not clear.
In this study, we demonstrate that Pro A inhibits PC cells at the nano-molar concentrations by inducing apoptosis and autophagy. Endogenous SMAD4 expression is associated with the sensitivity of PC cells to Pro A, while Pro A reduces the stability of SMAD4 protein. Our findings suggest that Pro A is a promising candidate for treating certain types of PC. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1085/rc).
Methods
Cell culture and drug treatment
PC cell lines (Panc-1, AsPC-1, BxPC-3) were from the Cell Bank of Shanghai Institute of Biological Sciences, China Academy of Sciences. All cell lines were cultured in RPMI 1640 medium (Corning, New York, NY, USA) at 37 ℃ with 5% CO2. Panc-1 cell cultures were supplemented with 10% fetal bovine serum (GIBCO, New York, NY, USA), while AsPC-1 and BxPC-3 cell cultures were supplemented with 20% fetal bovine serum. A master solution (10 mM) of Pro A (Sigma Aldrich, New York, NY, USA, #R214310) was prepared by dissolving Pro A in dimethyl sulfoxide (DMSO) and stored at −80 ℃. For mouse treatment, Pro A was first dissolved in DMSO at 50 mg/mL and then diluted with phosphate buffer saline (PBS) to a final concentration of 0.5 mg/mL before use. The solution was briefly heated at 42 ℃ and sonicated till clear.
Transfection, lentivirus construction and transduction
pCDH-SMAD4 was generated by subcloning SMAD4 cDNA amplified from pcDNA3.1-3xflag-SMAD4 (Youbio, Changsha, China) in pCDH (Addgene, New York, NY, USA). The coding DNA for the short hairpin RNA (shRNA) precursor targeting SMAD4 was inserted into pLKO.1 (Addgene), and co-transfected with packaging plasmids psPAX2 and pMD2.G (Addgene) into HEK293T cells for production of recombinant lentivirus. Transfections were performed using TurboFect Transfection Reagent (Thermo Fisher Scientific, Shanghai, China) following the manufacturer’s instructions. The shRNA sequences are as follows: shSMAD4-1: 5'-CCGGGGTGTGCAGTTGGAATGTACTCGAGTACATTCCAACTGCACACCTTTTTG-3'; shSMAD4-2: 5'-CCGGGTAGGACTGCACCATACACCTGCAGGTGTATGGTGCAGTCCTACTTTTTG-3'; shSMAD4-3: 5'-CCGGGTAGGACTGCACCATACACCTGCAGGTGTATGGTGCAGTCCTACTTTTTG-3'; shCtrl: 5'-CCGGAAAATGCAGTTGGAATGTACTCGAGTACATTCCAACTGCATTTTTTTTTG-3'. PC cells were transduced by recombinant lentivirus in the presence of 2 µg/mL polybrene (Sigma-Aldrich). The expression of target genes was verified with western blot.
Colony formation assay
PC cells were inoculated in triplicates in 6-well plates (500 cells/well for Panc-1 and BxPC-3, 1,000 cells/well for AsPC-1), and treated with different concentrations of Pro A. Cells were left to form colonies at 37 ℃ for 14 days. The cell colonies were washed and fixed with 4% paraformaldehyde (PFA) for 15 min and stained with crystal violet for 20 min at room temperature. The colonies were recorded under a microscope and analyzed by ImageJ.
Cell migration and invasion assays
In cell migration assays using a 24-well transwell plate, PC cells were treated with different concentrations of Pro A for 24 h and then suspended in serum-free medium. The 100 µL of suspensions was added on top of the transwell membrane and 600 µL of serum-containing medium was added in the lower chamber. After 48 h, cells on the membrane were fixed with 4% PFA and stained with 0.1% crystal violet. In cell invasion assays, the extracellular matrix (Matrigel, BD Biosciences, New York, NY, USA) was added on top of the transwell membrane. The following steps were the same as in migration assays. Cells were examined under an inverted microscope and the average number of cells in five randomly selected areas were calculated.
The directional cell migration was examined using wound-healing assay. After Pro A treatment for 12 h, cells were inoculated into 6-well plates. When the cell confluence reached 80–90%, the culture medium was replaced with serum-free medium. A pipette tip was used to make a straight scratch. At 0, 6, 12, 24 and 36 h after scratch, the active edge of the cells was observed under a microscope and photographed.
Cell proliferation assay
The inhibitory effect of Pro A on cell proliferation was determined by cell counting kit-8 (CCK-8) (Dojindo, Kumamoto, Japan). Cells were inoculated in 96-well plates (6,000 cells/well for Panc-1 and BxPC-3, 7,000 cells/well for AsPC-1). The medium was refreshed 12 h after inoculation and cells were treated with increasing concentrations of Pro A with 5 wells for each concentration and cultured for 72 h. CCK-8 reagent was added and OD450 values were measured with Epoch Microplate Spectrophotometer (Bio-Tek, New York, NY, USA). Data were plotted and half-maximal inhibitory concentration (IC50) was calculated using Graphpad 7.
Cell cycle analysis and apoptosis assays
PC cells treated with Pro A for 48-h were fixed overnight with 70% ethanol, stained for 30 min at 37 ℃ in dark with propidium iodide (PI; Beyotime Biotechnology, Shanghai, China) for cell cycle analysis or with Annexin V-FITC Apoptosis Detection Kit (Dojindo) for apoptosis assays. Cells were then analyzed using FACSCalibur flow cytometry (BD Biosciences).
Reactive oxygen species (ROS) measurement
Cells were disassociated through trypsin treatment, washed three times with PBS, and then incubated with 10 µM 2’,7’-dichlorofluorescein diacetate (DCFH-DA; Yeasen, Shanghai, China) for 30 min at 37 ℃ in dark. After the incubation, cells were washed and collected. Fluorescence was measured using FACScalibur flow cytometry (BD Biosciences) at 488 nm (excitation) and 525 nm (emission).
Determination of intracellular free Ca2+
Pro A-treated PC cells were washed with PBS, resuspended in serum-free medium containing 4 µM Fluo-4 AM (Yeasen), and incubated for 30 min at 37 ℃ in dark. Cells were then washed with PBS and fluorescence was measured using FACScalibur flow cytometry (BD Biosciences) at 494 nm (excitation) and 516 nm (emission).
Determination of mitochondrial membrane potential (MMP)
Pro A-treated PC cells were washed with PBS and stained with 10 µg/mL JC-1 fluorescent probe (Yeasen) for 15 min at 37 ℃ in dark. Cells were then washed with PBS, and green and red fluorescence, which represent JC-1 monomer and aggregates respectively, was measured using FACSCalibur flow cytometry (BD Biosciences). MMP was calculated by a decrease in red/green fluorescence intensity ratio.
Western blot
Protein samples were mixed with 5× SDS loading buffer (Beyotime Biotechnology) and heated at 100 ℃ in metal bath. After thorough separation, proteins were transferred onto nitrocellulose membrane (Millipore, New York, NY, USA) using Trans-blot Turbo system (Bio-Rad, Hercules, CA, USA). The membrane was blocked with PBST containing 3% bovine serum albumin (Sangon Biotech, Shanghai, China) and then incubated with a specific primary antibody overnight at 4 ℃. Antibodies for PARP, cleaved-PARP, Caspase 9, Caspase 3, cleaved-Caspase 3, P62, LC3, mTOR, Phospho-mTOR, Raptor, ATG7, SMAD4, E-cadherin and ZO-1 were from Cell Signaling Technology (Boston, MA, USA). Antibodies for MMP2 were from Servicebio (Shanghai, China). Signals were detected using ECL substrate (Millipore) and captured using ChemiDoc XRS+ (Bio-Rad).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted with TRIzol Reagent (Invitrogen, Los Angeles, CA, USA). cDNA was synthesized by reverse transcription using Prime Script RT reagent Kit (TaKaRa, Kyoto, Japan) according to the manufacturer’s instructions. Quantitative real-time PCR was performed with SYBR Green SuperReal Premix (TIANGEN, Beijing, China) on CFX Connect Real-Time System (Bio-Rad). The expression of target genes was determined using the 2−ΔΔCt method and normalized to that of the housekeeping gene GAPDH. Primer sequences are shown in Table S1.
GFP-LC3 fluorescence microscopy
Cells grown on microscope cover glasses (Jingan Biological, Shanghai, China) in a 12-well plate were transfected with pGFP-LC3 (a gift from Dr. Qiang Deng of Shanghai Medical College). Six hours after transfection, cells were treated with Pro A for 24 h. Cells were then fixed with 4% PFA, treated with 0.2% Triton X-100 and stained with 4’,6-diamidino-2’-phenylindole (DAPI). Cell images were obtained using a laser scanning confocal microscope (Leica, Heidelberg, Germany).
Tumor xenograft mouse model
Four-week-old female BALB/c nude mice weighing about 16 g were from Shanghai Jihui Laboratory Animal Care and housed in Fudan experimental SPF animal center. Seven days after subcutaneous implantation of 5×106 Panc-1 cells into the left armpit, 12 mice were randomly divided into two groups and injected with vehicle (PBS) or Pro A (6.5 mg/kg) every two days for 3 weeks before sacrifice. Tumor volumes were measured every three days. Experiments were performed under a project license (No. 202103005S) granted by the Animal Ethics Committee of School of Basic Medical Sciences, Fudan University and were performed in accordance with the guidelines of the National Institutes of Health on the care and use of laboratory animals.
Statistical analysis
Data were analyzed using Graphpad Prism 7 and expressed as mean ± standard error of mean (SEM). Student t-test and one-way analysis of variance (ANOVA) were used to compare two groups and multiple groups, respectively. P<0.05 was considered statistically significant.
Results
Pro A inhibits proliferation, migration and invasion of PC cell lines in vitro
To investigate the anti-tumor activity of Pro A on PC, we first examined the cytotoxic effect of Pro A on three PC cell lines Panc-1, BxPC-3 and AsPC-1. A 72-h treatment of Pro A inhibited cell proliferation of all three cell lines in a dose-dependent manner. The half-maximal inhibitory concentration (IC50) values for Panc-1, BxPC-3 and AsPC-1 were 35.25, 180.3 and 370.9 nM, respectively (Figure 1A, Table 1). Cell proliferation decreased with the extension of treatment, showing that the inhibitory effect was also time-dependent (Figure 1B). Notably, Panc-1 cells were more sensitive to Pro A than BxPC-3 and AsPC-1 cells. The inhibitory effect of Pro A was further confirmed by colony formation assays (Figure 1C, Table 1) and cell morphology (Figure S1). These results indicated that Pro A inhibits the proliferation of PC cells and different PC cell lines display different sensitivity to Pro A.
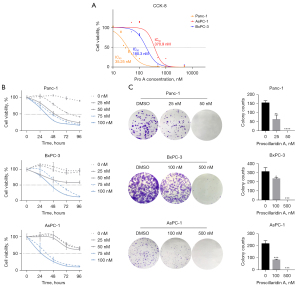
Table 1
| Pro A on cell line | Panc-1 [IC50 (CCK-8) =35.25 nM] | BxPC-3 [IC50 (CCK-8) =180.3 nM] | AsPC-1 [IC50 (CCK-8) =370.9 nM] | |||||
|---|---|---|---|---|---|---|---|---|
| 25 nM | 50 nM | 100 nM | 500 nM | 100 nM | 500 nM | |||
| Colony formation | 41.1%** | 0%**** | 75.2%* | 0%*** | 38.3%*** | 0%*** | ||
| Migration | 60.3%**** | 9.5%**** | 13.3%**** | 0%**** | 44.5%**** | 0%**** | ||
| Invasion | 17.5%**** | 0%**** | 80.7% | 15.6%**** | 81.1%*** | 25.6%**** | ||
| Apoptosis | 122.0% | 213.3%*** | 134.3% | 524.3%**** | 93.5% | 192.1%** | ||
| ROS | 140.6% | 175.3%* | 202.3%**** | 285.2%**** | 195.1% | 577.2%**** | ||
| Ca2+ | 100.4% | 130.5%** | 227.6%*** | 421.4%**** | 79.3%* | 204.4%**** | ||
| MMP | 42.0%* | 30.9%* | 127.7% | 22.0%* | 94.4% | 12.2%*** | ||
Results presented in figures are summarized here. Percentages are calculated by normalizing data from Pro A-treated cells against DMSO-treated cells. Statistical significance was calculated using Student t-test. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. Pro A, proscillaridin A; PC, pancreatic cancer; CCK-8, cell counting kit-8; ROS, reactive oxygen species; MMP, mitochondrial membrane potential; DMSO, dimethyl sulfoxide.
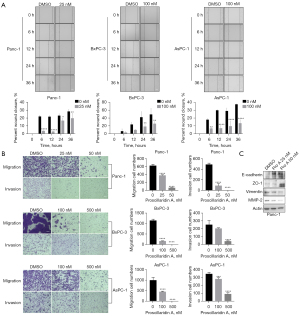
The effects of Pro A on migration and invasion capacities of PC cell lines were tested using transwell and wound-healing assays. Migration and invasion were significantly reduced after Pro A treatment (Figure 2A,2B, Table 1). Epithelial-mesenchymal transition (EMT), a process in which epithelial cells acquire mesenchymal features, is associated with tumor initiation, invasion, metastasis, and resistance to therapy. We detected the key molecular markers of EMT by western blot to determine whether Pro A affects the EMT process. Pro A treatment of PC cells resulted in decreased expression of cell invasion and metastasis markers Vimentin and MMP2, and increased expression of markers of suppression of cell invasion E-cadherin and ZO-1 (Figure 2C). These data suggested that Pro A reduces the metastatic potential of the PC cells.
Pro A inhibits tumor growth in Panc-1-xenografted mice
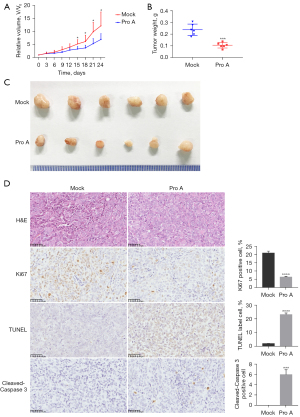
To evaluate the anti-tumor effect of Pro A on tumor growth in vivo, Panc-1 cells were subcutaneously injected into the left armpit of nude mice (5×106 cells/mouse). Mice were randomly divided into two groups, receiving either 6.5 mg/kg of Pro A or the same volume of PBS every 2 days. Tumor volumes were measured every 3 days. The results showed that starting from day 15 post-treatment, the mean tumor volume of Pro A-treated mice was significantly smaller than the PBS group (Figure 3A). On day 24, mice were sacrificed and the mean tumor weight of Pro A-treated mice was markedly lower than PBS group (0.11±0.02 vs. 0.24±0.05 g, P<0.0001, n=6) (Figure 3B,3C). The assays of TUNEL, Ki67 and cleaved-Caspase 3 can indicate cell proliferation and apoptosis, respectively. Immunohistochemical results showed that compared with PBS treated mice, tumors in Pro A-treated mice contained fewer Ki67 positive cells and more cleaved-Caspase 3 positive cells (Figure 3D). These results indicate that Pro A inhibits PC progression in xenografted mice.
Pro A induces apoptosis and blocked cell cycle progression
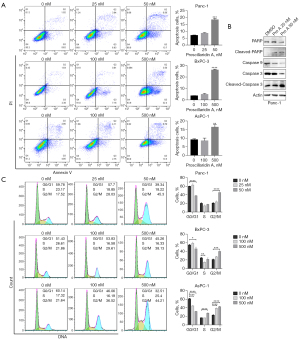
Compared with control cells, PC cells treated with Pro A for 48 h showed significantly increased apoptosis as assayed by an annexin V/PI staining. Similarly, Panc-1 cells were more sensitive to Pro A induced apoptosis than BxPC-3 and AsPC-1 cells (Figure 4A, Table 1). In Panc-1 cells treated with Pro A for 48 h, molecular markers of apoptosis (decreased levels of caspase 9 and increased levels of cleaved caspase 3 and PARP) could be easily observed (Figure 4B). We further examined the effect of Pro A on PC cell cycle progression. The results showed that Pro A treatment resulted in the increase of G2 phase PC cells and the decrease of S phase PC cells (Figure 4C). These results suggest that Pro A can induce PC cell apoptosis and block the progression of cell cycle.
Pro A induces mitochondrial damage in PC cells
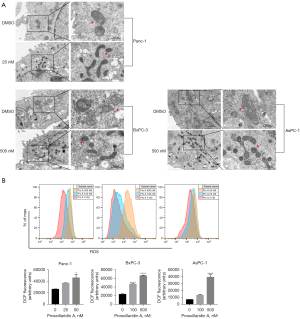
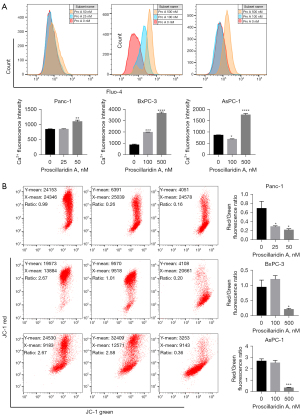
Mitochondria are the site of adenosine triphosphate (ATP) synthesis. Mitochondrial damage is characterized by a series of changes, including cytoplasmic Ca2+ accumulation, ROS production and MMP decrease (26). We first examined mitochondria in PC cells treated with Pro A by transmission electron microscopy. In Pro A-treated PC cells, mitochondria became long rod-shaped and swollen (Figure 5A). DCFH-DA staining showed that ROS levels increased significantly in PC cells treated with Pro A for 48 h and in a dose-dependent manner (Figure 5B, Table 1). Intracellular Ca2+ levels, assayed using fluo-4-Ca2+ imaging, also increased with Pro A treatment (Figure 6A, Table 1). MMP in PC cells was analyzed using JC-1 staining, which showed significant decrease in MMP after Pro A treatment (Figure 6B, Table 1). The data showed that Pro A induced mitochondrial damage.
Pro A induces autophagy in PC cells
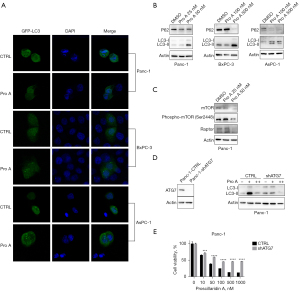
Autophagy is a cellular mechanism for coping with stress, defense and recycling. When autophagy occurs, the cleavage of LC3 by ATG4 produces the soluble form LC3-I, which is then bound to the membrane of emerging autophagosomes by ATG3 and ATG7, where LC3-I is transformed to the lipidated form LC3-II.
To determine the role of Pro A in autophagy activation, we used the GFP-LC3-fluorescent indicator system (27). PC cells were transfected with pGFP-LC3 and treated with Pro A for 24 h. GFP-LC3 aggregation and increased LC3-II levels were observed in Pro A-treated PC cells by a fluorescent microscopy and western blot, respectively (Figure 7A,7B). mTOR and Raptor, which inhibit autophagy, were down-regulated after 50 nM Pro A treatment in Panc-1 cells (Figure 7C). To assess whether autophagy affects Pro A-mediated cytotoxicity, shATG7 was stably transfected into Panc-1 cells to reduce the expression of ATG7. Knockdown of ATG7 expression inhibited the production of LC3-II and the cytotoxicity of Pro A was partially relieved (Figure 7D,7E), suggesting that Pro A partially reduces PC cell viability through induction of autophagy.
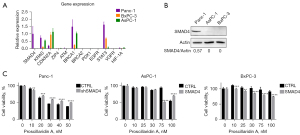
SMAD4 expression partially determines the sensitivity of PC cells to Pro A
To explore why Panc-1 is more sensitive to Pro A treatment than BxPC-3 and AsPC-1, we used qPCR to detect the expression of important cancer-related genes in the three PC cell lines (Figure 8A). Panc-1 cells had significantly higher SMAD4 mRNA expression than BxPC-3 and AsPC-1 cells, which was confirmed at the protein level (Figure 8B). In order to further study the relationship between Pro A’s anti-tumor activity and SMAD4 expression, we constructed three stably transfected cell lines, in which SMAD4 protein was down-regulated in Panc-1 and overexpressed in BxPC-3 and AsPC-1 (Figure S2). The cytotoxicity of Pro A was slightly alleviated with SMAD4 knockdown in Panc-1 cells, and became slightly aggravated with SMAD4 overexpressed in BxPC-3 and AsPC-1 cells (Figure 8C). The results showed that SMAD4 expression may be related to the cytotoxicity of Pro A for PC cells.
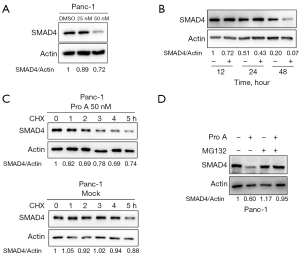
Interestingly, Pro A treatment also reduced endogenous SMAD4 protein levels in Panc-1 cells in a dose-dependent and time-dependent manner (Figure 9A,9B). To understand how Pro A reduces SMAD4 protein expression, we treated Panc-1 cells with cycloheximide (CHX) and measured protein level of SMAD4. The results showed that the SMAD4 protein levels decreased faster in the presence of Pro A, which indicates that Pro A might affect the degradation of SMAD4 protein (Figure 9C). Consistent with this observation, SMAD4 protein levels in Panc-1 cells treated with proteasome inhibitor MG132 were almost unaffected by Pro A (Figure 9D). These results suggest that Pro A might promote SMAD4 degradation through the ubiquitination-proteasome pathway.
Discussion
CGs are a class of natural products used traditionally to increase cardiac contractility in patients with congestive heart failure and arrhythmia by blocking the Na+/K+ ATPase of the heart. Previous studies have reported that CGs can induce cell death of senescent cells and increase the levels of intracellular Ca2+ and H+ to cause anti-tumor effects (28-30). Pro A belongs to the CG family and comes from Drimia maritima. Its therapeutic efficacy was first described more than two centuries ago (31). Although it is no longer used clinically, Pro A was reported to have anti-tumor effects on several cancers. Pro A can inhibit STAT3 signaling in breast cancer cells (32) and trigger the activation of endoplasmic reticulum stress to slow the prostate cancer progression (33). Previous study has shown that Pro A at micro-molar concentrations can induce PC cell apoptosis (25). In this study, we found that Pro A at the nano-molar concentrations has cytotoxic effects on PC cells, including inhibiting the proliferation, migration and invasion of PC cells in vitro (Figures 1,2) and tumor growth in mouse models (Figure 3). In addition, Pro A induces cell death by promoting both apoptosis (Figures 4-6) and autophagy (Figure 7). It is worth noting that Panc-1, BxPC-3 and AsPC-1 cells react differently to the cytotoxicity of Pro A, with Panc-1 being much more sensitive to Pro A (Table 1), which may be partly due to SMAD4 expression in Panc-1 (Figures 8,9).
SMAD4, which serves as the central mediator of TGF-β signaling, translocates to the nucleus as SMAD2/SMAD3-SMAD4 complex after TGF-β family receptor activation. SMAD4 and related pathway components are inactivated in ~50% of PC and cooperate with activated KrasG12D to promote PDAC in mouse model (34), but the role of SMAD4-mediated pathway in PC progression has not been elucidated. Cell proliferation was reported to be increased in SMAD4-expressing Panc-1 cells and decreased in SMAD4-null BxPC-3 cells upon TGFβ-1 stimulation (35). However, another report suggested that inactivation of TGF-β signaling in a mouse model enhanced PC initiation and progression in a SMAD4-dependant manner (36). In this work, Panc-1 displayed the highest sensitivity to Pro A among the three tested PC cell lines (Figure 1A). Previous genotyping reports showed that SMAD4, also named DPC4 for deleted in pancreatic carcinoma locus 4, is un-mutated in Panc-1, but lost due to homozygous deletion in BxPC-3 (37-39). There have been conflicting reports regarding the status of SMAD4 in AsPC-1 (40,41). Our data from mRNA and protein analyses show that Panc-1 was indeed the only PC cell line tested that displayed endogenous SMAD4 expression, which was related to sensitivity to Pro A, with downregulation of SMAD4 expression in Panc-1 dampening Pro A’s anti-tumor effect (Figure 8C). Furthermore, we showed that Pro A reduced the stability of SMAD4 (Figure 9C,9D). The wild-type SMAD4 has been shown to mainly undergo mono- and oligo-ubiquitination, which enhances its interaction with other SMADs and results in higher transcriptional activation, instead of poly-ubiquitination, which leads to degradation through proteasome (42). It is plausible that Pro A treatment, through as yet unknown mechanisms, promotes the poly-ubiquitination of SMAD4. On the other hand, there was no significant difference in the cell proliferation between the wild-type and SMAD4 knockdown Panc-1 cells (Figure S3). These data suggest that SMAD4 expression partially determines the sensitivity of PC cells to Pro A. How SMAD4 impacts on Pro A’s anti-tumor activity is currently unclear and warrants further study.
It is widely accepted that mitochondria play a crucial role in cell proliferation and apoptosis. Caspase family proteins and PARP proteins are considered to be the main participants in mitochondrial apoptosis. We found that the expression of cleaved caspase-3 and PARP increased in Pro A-treated PC cells (Figure 4B). The production of ROS, the disruption of MMP and the increase in intracellular Ca2+ level are the main characteristics of endogenous or mitochondrial apoptosis, which all manifested in PC cells treated with Pro A (Figures 5,6). The results confirm the involvement of mitochondrial apoptosis in Pro A’s anti-tumor activity against PC cells. It’s worth noting that intracellular Ca2+ levels in Panc-1 cells treated with cytotoxic concentrations of Pro A increased only moderately compared to the other two cell lines (Figure 6A) and this apparent discordance is more likely a result of differences between these cells in other mechanisms affecting Ca2+ levels, for instance endoplasmic reticulum (ER) sequestration and plasma membrane Ca2+ pump(s). Moreover, it is indeed possible that different sensitivities to Pro A between the three PC cell lines used here might also be related to differences in Na+/K+ ATPase expression, its pump activity and downstream signaling.
Furthermore, our data demonstrate that Pro A not only induces apoptosis, but also induces a certain degree of autophagy. Autophagy is a major intracellular degradation system, which has opposing, context-dependent roles in cancers. Both stimulation and inhibition of autophagy have been proposed as cancer treatment (43). CGs have been shown to induce autophagy in some cancer types (44-46). Here we show for the first time that Pro A induces autophagy in PC cells, characterized by the changes in the expression of autophagy markers, LC3-I/II, p62 and mTOR pathway (Figure 7). In addition, ATG7-deficient Panc-1 cells showed reduced sensitivity to Pro A treatment. These data suggest that autophagy may also contribute to the anti-tumor activity of Pro A on PC cells.
In conclusion, our findings provide insights into the anti-tumor effect of Pro A on PC cells and a starting point on elucidation of the molecular mechanism of Pro A’s differential cytotoxic effects on different PC cells, which will lay the basis for Pro A and similar molecules to be tested as novel drugs for PC treatment. More in-depth work is required to identify additional traits associated with sensitivity to Pro A, preferably using primary PC tumor cells, which would hopefully lead to the establishment of a practical subtyping criteria for screening candidates for initial testing of Pro A in human PC.
Acknowledgments
Funding: This study was supported by the NSFC (No. 81971921), National Key Project for Infectious Diseases of China (Nos. 2017ZX10202202 and 2018ZX10301208), and Shanghai Municipal Education Commission (No. 201701070007E00057).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1085/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1085/dss
Peer Review File: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1085/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1085/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (No. 202103005S) granted by the Animal Ethics Committee of School of Basic Medical Sciences, Fudan University and were performed in accordance with the guidelines of the National Institutes of Health on the care and use of laboratory animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Grasso C, Jansen G, Giovannetti E. Drug resistance in pancreatic cancer: Impact of altered energy metabolism. Crit Rev Oncol Hematol 2017;114:139-52. [Crossref] [PubMed]
- Klein AP. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat Rev Gastroenterol Hepatol 2021;18:493-502. [Crossref] [PubMed]
- Naudin S, Li K, Jaouen T, et al. Lifetime and baseline alcohol intakes and risk of pancreatic cancer in the European Prospective Investigation into Cancer and Nutrition study. Int J Cancer 2018;143:801-12. [Crossref] [PubMed]
- Amaral AF, Porta M, Silverman DT, et al. Pancreatic cancer risk and levels of trace elements. Gut 2012;61:1583-8. [Crossref] [PubMed]
- Antwi SO, Eckert EC, Sabaque CV, et al. Exposure to environmental chemicals and heavy metals, and risk of pancreatic cancer. Cancer Causes Control 2015;26:1583-91. [Crossref] [PubMed]
- The Lancet Gastroenterology Hepatology. Pancreatic cancer: a state of emergency? Lancet Gastroenterol Hepatol 2021;6:81. [Crossref]
- Chiorean EG, Coveler AL. Pancreatic cancer: optimizing treatment options, new, and emerging targeted therapies. Drug Des Devel Ther 2015;9:3529-45. [Crossref] [PubMed]
- Neoptolemos JP, Kleeff J, Michl P, et al. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol 2018;15:333-48. [Crossref] [PubMed]
- Ducreux M, Seufferlein T, Van Laethem JL, et al. Systemic treatment of pancreatic cancer revisited. Semin Oncol 2019;46:28-38. [Crossref] [PubMed]
- Wang Y, Yang G, You L, et al. Role of the microbiome in occurrence, development and treatment of pancreatic cancer. Mol Cancer 2019;18:173. [Crossref] [PubMed]
- Grant TJ, Hua K, Singh A. Molecular Pathogenesis of Pancreatic Cancer. Prog Mol Biol Transl Sci 2016;144:241-75. [Crossref] [PubMed]
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47-52. [Crossref] [PubMed]
- Mann KM, Ying H, Juan J, et al. KRAS-related proteins in pancreatic cancer. Pharmacol Ther 2016;168:29-42. [Crossref] [PubMed]
- Zhen DB, Rabe KG, Gallinger S, et al. BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic cancer: a PACGENE study. Genet Med 2015;17:569-77. [Crossref] [PubMed]
- Escobar-Hoyos LF, Penson A, Kannan R, et al. Altered RNA Splicing by Mutant p53 Activates Oncogenic RAS Signaling in Pancreatic Cancer. Cancer Cell 2020;38:198-211.e8. [Crossref] [PubMed]
- Wang Z, Li Y, Zhan S, et al. SMAD4 Y353C promotes the progression of PDAC. BMC Cancer 2019;19:1037. [Crossref] [PubMed]
- Duan Y, Yin X, Lai X, et al. Upregulation of DAB2IP Inhibits Ras Activity and Tumorigenesis in Human Pancreatic Cancer Cells. Technol Cancer Res Treat 2020;19:1533033819895494. [Crossref] [PubMed]
- Schneider NFZ, Cerella C, Simões CMO, et al. Anticancer and Immunogenic Properties of Cardiac Glycosides. Molecules 2017;22:1932. [Crossref] [PubMed]
- Cerella C, Dicato M, Diederich M. Assembling the puzzle of anti-cancer mechanisms triggered by cardiac glycosides. Mitochondrion 2013;13:225-34. [Crossref] [PubMed]
- Bielawski K, Winnicka K, Bielawska A. Inhibition of DNA topoisomerases I and II, and growth inhibition of breast cancer MCF-7 cells by ouabain, digoxin and proscillaridin A. Biol Pharm Bull 2006;29:1493-7. [Crossref] [PubMed]
- He Y, Khan M, Yang J, et al. Proscillaridin A induces apoptosis, inhibits STAT3 activation and augments doxorubicin toxicity in prostate cancer cells. Int J Med Sci 2018;15:832-9. [Crossref] [PubMed]
- Luo M, Liu Y, Liu N, et al. Proscillaridin A inhibits hepatocellular carcinoma progression through inducing mitochondrial damage and autophagy. Acta Biochim Biophys Sin (Shanghai) 2021;53:19-28. [Crossref] [PubMed]
- Da Costa EM, Armaos G, McInnes G, et al. Heart failure drug proscillaridin A targets MYC overexpressing leukemia through global loss of lysine acetylation. J Exp Clin Cancer Res 2019;38:251. [Crossref] [PubMed]
- Li RZ, Fan XX, Duan FG, et al. Proscillaridin A induces apoptosis and suppresses non-small-cell lung cancer tumor growth via calcium-induced DR4 upregulation. Cell Death Dis 2018;9:696. [Crossref] [PubMed]
- Kota S, Hou S, Guerrant W, et al. A novel three-dimensional high-throughput screening approach identifies inducers of a mutant KRAS selective lethal phenotype. Oncogene 2018;37:4372-84. [Crossref] [PubMed]
- Yang Y, Karakhanova S, Hartwig W, et al. Mitochondria and Mitochondrial ROS in Cancer: Novel Targets for Anticancer Therapy. J Cell Physiol 2016;231:2570-81. [Crossref] [PubMed]
- Masud S, Prajsnar TK, Torraca V, et al. Macrophages target Salmonella by Lc3-associated phagocytosis in a systemic infection model. Autophagy 2019;15:796-812. [Crossref] [PubMed]
- Olona A, Hateley C, Guerrero A, et al. Cardiac glycosides cause cytotoxicity in human macrophages and ameliorate white adipose tissue homeostasis. Br J Pharmacol 2022;179:1874-86. [Crossref] [PubMed]
- López-Lázaro M. Digitoxin as an anticancer agent with selectivity for cancer cells: possible mechanisms involved. Expert Opin Ther Targets 2007;11:1043-53. [Crossref] [PubMed]
- Xie CM, Chan WY, Yu S, et al. Bufalin induces autophagy-mediated cell death in human colon cancer cells through reactive oxygen species generation and JNK activation. Free Radic Biol Med 2011;51:1365-75. [Crossref] [PubMed]
- Triana-Martínez F, Picallos-Rabina P, Da Silva-Álvarez S, et al. Identification and characterization of Cardiac Glycosides as senolytic compounds. Nat Commun 2019;10:4731. [Crossref] [PubMed]
- Saleem MZ, Alshwmi M, Zhang H, et al. Inhibition of JNK-Mediated Autophagy Promotes Proscillaridin A- Induced Apoptosis via ROS Generation, Intracellular Ca+2 Oscillation and Inhibiting STAT3 Signaling in Breast Cancer Cells. Front Pharmacol 2020;11:01055. [Crossref] [PubMed]
- Wang F, Liu L, Tong Y, et al. Proscillaridin A slows the prostate cancer progression through triggering the activation of endoplasmic reticulum stress. Cell Cycle 2020;19:541-50. [Crossref] [PubMed]
- Zhao M, Mishra L, Deng CX. The role of TGF-β/SMAD4 signaling in cancer. Int J Biol Sci 2018;14:111-23. [Crossref] [PubMed]
- Malinda RR, Zeeberg K, Sharku PC, et al. TGFβ Signaling Increases Net Acid Extrusion, Proliferation and Invasion in Panc-1 Pancreatic Cancer Cells: SMAD4 Dependence and Link to Merlin/NF2 Signaling. Front Oncol 2020;10:687. [Crossref] [PubMed]
- Hezel AF, Deshpande V, Zimmerman SM, et al. TGF-β and αvβ6 integrin act in a common pathway to suppress pancreatic cancer progression. Cancer Res 2012;72:4840-5. [Crossref] [PubMed]
- Deer EL, González-Hernández J, Coursen JD, et al. Phenotype and genotype of pancreatic cancer cell lines. Pancreas 2010;39:425-35. [Crossref] [PubMed]
- Loukopoulos P, Kanetaka K, Takamura M, et al. Orthotopic transplantation models of pancreatic adenocarcinoma derived from cell lines and primary tumors and displaying varying metastatic activity. Pancreas 2004;29:193-203. [Crossref] [PubMed]
- Aoki Y, Hosaka S, Tachibana N, et al. Reassessment of K-ras mutations at codon 12 by direct PCR and sequencing from tissue microdissection in human pancreatic adenocarcinomas. Pancreas 2000;21:152-7. [Crossref] [PubMed]
- Moore PS, Sipos B, Orlandini S, et al. Genetic profile of 22 pancreatic carcinoma cell lines. Analysis of K-ras, p53, p16 and DPC4/Smad4. Virchows Arch 2001;439:798-802. [Crossref] [PubMed]
- Sun C, Yamato T, Furukawa T, et al. Characterization of the mutations of the K-ras, p53, p16, and SMAD4 genes in 15 human pancreatic cancer cell lines. Oncol Rep 2001;8:89-92. [Crossref] [PubMed]
- Morén A, Hellman U, Inada Y, et al. Differential ubiquitination defines the functional status of the tumor suppressor Smad4. J Biol Chem 2003;278:33571-82. [Crossref] [PubMed]
- Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer 2017;17:528-42. [Crossref] [PubMed]
- Škubník J, Svobodová Pavlíčková V, Psotová J, et al. Cardiac Glycosides as Autophagy Modulators. Cells 2021;10:3341. [Crossref] [PubMed]
- Trenti A, Grumati P, Cusinato F, et al. Cardiac glycoside ouabain induces autophagic cell death in non-small cell lung cancer cells via a JNK-dependent decrease of Bcl-2. Biochem Pharmacol 2014;89:197-209. [Crossref] [PubMed]
- Wang Y, Qiu Q, Shen JJ, et al. Cardiac glycosides induce autophagy in human non-small cell lung cancer cells through regulation of dual signaling pathways. Int J Biochem Cell Biol 2012;44:1813-24. [Crossref] [PubMed]


