
Pluripotency of periodontal ligament and dental pulp cells is induced by intercellular communication via the CDX2/Oct-4/Sox2 pathway
Introduction
The renewal of dental tissue has long posed a significant challenge. Many treatments have been attempted, including tissue transplantation and the use of biomaterials and growth factors; however, none have proved satisfactory (1-3). The use of cell-based treatments has been recently proposed and has provided insights into the processes of dental tissue regenerative mechanisms and the potential for effective therapy (4,5). Periodontal ligament cells (PDLCs) and dental pulp cells (DPCs) are pluripotent and can differentiate into different cell types; this property has the potential to facilitate the repair and regeneration of dental tissue (6). Both cell types can be easily and non-invasively harvested during normal dental procedures (7) and retains the potential to differentiate into dental chondrocytes, osteoblasts and adipocytes (8,9). Despite inevitable senescence in culture, resulting in potential phenotypic alterations, the cells represent promising repositories of stem cells for repairing dental tissue (10). Thus, the maintenance of the stemness of DPCs and PLDCs in culture is an important challenge.
Epigenetic regulation plays a crucial role in the renewal of tooth tissue, such as increased levels of acetylated histone H3 lysine 9 (H3K9ac) and H3K27ac involved in odontoblast differentiation (11). Caudal-type homeobox transcription factor 2 (CDX2) is a transcription factor that has been shown to modulate proliferation and differentiation in the intestine (12). Recent research has shown that CDX2 also functions as an oncogene and tumor suppressor, as well as a regulator of cellular reprogramming and pluripotency during embryonic development (13,14). However, it is not yet known whether CDX2 regulates the pluripotency of human DPCs and PDLCs.
Intercellular communication in the culture environment can stimulate the expression of pluripotency markers in differentiated cells (15). Cells can be maintained in an undifferentiated state in culture by the addition of specific factors or the use of feeder cells (16-18). The transfection of mouse fibroblasts with Sox2, Oct-4 and MYC proto-oncogene, bHLH transcription factor (c-Myc) has been shown to induce qualities of pluripotency similar to those of embryonic stem cells (19,20). The endogenous expression levels of these genes have also been shown to correlate with pluripotency in dental cells with Oct-4 and Sox2 acting in concert (21,22). The influence of the culture environment on pluripotency is also well documented, and the inclusion of growth factors restore and regenerate aged and mature cells and stimulate the expression of pluripotency-associated genes. Co-cultures of DPCs with endothelial cells may also promote osteogenic/odontogenic differentiation (23). These findings emphasize the significance of signaling pathways in the regulation of intercellular communication.
In this study, we assessed the expression of pluripotency markers to explore the cell-to-cell interactions regulating specific signaling pathways and pluripotency-associated genes in dental cells using indirect co-culture systems. It is hoped that our study of the pluripotency of PDLC and DPC will provide new insights into the regeneration of tooth tissue repair. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3492/rc).
Methods
Culture of DPCs and PDLCs
PDLCs and DPCs were obtained from the extracted molars of young patients. The explant cultures were maintained as described previously (24). Third-generation cells were used for the experiments. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Sun Yat-sen University (No. 2018-02-142) and informed consent was taken from all the patients.
Differential expression analysis of stem cell-related genes
Total RNA was extracted from cells using TRIzol (Invitrogen). Differentially expressed genes (DEGs), which were defined as genes showing a minimum 3-fold change in expression in comparison to the control genes (P<0.05), were identified by a stem cell-associated RT2 Profiler PCR array in accordance with the provided protocol. The DEGs were assigned to groups according to their function. Protein levels were evaluated by western blotting.
Transfection of CDX2 in DPCs
Human CDX2 gene overexpression plasmid pcDNA4.0-CDX2, and short-hair RNA plasmids were obtained from GenePharma (Guangzhou, China). DPCs and PDLCs (104/mL) were plated in 24-well plates and grown for 24 h. The transfection of the plasmids was performed with Lipofectamine 3000 in accordance with the provided protocol. Messenger RNA (mRNA) and protein expression were determined after 48 h; The mRNA expression of CDX2, Oct-4 and Sox2 was assessed by RT-qPCR and protein expression was assessed by western blot. Cell-cycle and apoptosis analyses were performed by flow cytometry.
Quantitative real-time polymerase chain reaction (RT-qPCR)
Total RNA was extracted from DPCs or DPLCs using TRIzol reagent (Invitrogen). The RNA is then reverse transcribed to cDNA using the TaqMan™ Advanced miRNA cDNA Synthesis Kit or the First Strand cDNA Synthesis Kit. Oct-4 and Sox2 mRNA levels were assessed on day 5. The primers are listed in Table 1.
Table 1
| Gene | Primers or probes |
|---|---|
| Oct-4 (qPCR) | Forward: 5'-GCTCGAGAAGGATGTGGTC-3' |
| Reverse: 5'-ATCCTCTCGTTGTGCATAGTCG-3' | |
| Sox2 (qPCR) | Forward: 5'-GAGAACCCCAAGATGCACAAC-3' |
| Reverse: 5'-CGCTTAGCCTCGTCGATGA-3' | |
| CDX2 (qPCR) | Forward: 5'-CTGGTTTCAGAACCGCAGAG-3' |
| Reverse: 5'-GAAGACACCGGACTCAAGGG-3' | |
| Oct-4 (ChIP) | Forward: 5'-CATTGTACTCCACTGCACTCC-3' |
| Reverse: 5'-CCAGGACCTCAGTGCAGGTCC-3' | |
| Sox2 (ChIP) | Forward: 5'-GTTAGTAAGGAACAAAACAATG-3' |
| Reverse: 5'-CCTAAGAAAATGTACTGAATAG-3' | |
| Probe for Oct-4 | Probe: 5'-GTCTTAAAAAATAAAAATAAAAAAAGTTTCTGTGGG-3' |
| Probe for Sox2 | Probe: 5'-CCGTTTTGTAAAGATAATAAATGGAACGTGGCTGGTAG-3' |
Oct-4, octamer-binding transcription factor 4; Sox2, SRY-box transcription factor 2; CDX2, caudal-type homeobox transcription factor 2; ChIP, Chromatin Immunoprecipitation; EMSA, electrophoretic mobility shift assay; qPCR, quantitative polymerase chain reaction.
Western blotting
After cell lysis and clarification, the supernatant proteins (50 µg/lane) were electrophoresed and transferred to nitrocellulose. After blocking, the blots were probed with antibodies against CDX2 (1:2,000, Abcam, ab76541), Oct-4 (1:1,000, Abcam, ab181557), Sox2 (1:1,000, Abcam, ab92494), and GAPDH (1:1,000, Abcan, ab9485). After 1 h incubation with the corresponding secondary antibodies at room temperature, the bands were visualized by enhanced chemiluminescence.
Analysis of apoptosis and cell cycle
Flow cytometry was used to determine the cell-cycle phase and proportions of the apoptotic cells. For the former, cells in the lower chamber were harvested after 5 days in culture. Single-cell suspensions were treated with propidium iodide and applied to the flow cytometer. To measure apoptosis, cells in the lower chamber were collected after 5 days in culture, washed, and incubated with an Annexin V-Fluorescein 5-isothiocyanate (FITC)/propidium iodide (PI) solution (MultiSciences Biotech, Shanghai, China) in the dark before flow cytometric analysis. The apoptotic cells were Annexin V+/PI– and Annexin V+/PI+.
Dual-luciferase assays
The JASPAR CORE database was used to predict Oct-4 and Sox2 are potential targets of CDX2. Amplification of pGL3 plasmids containing wild-type (WT) or mutant (MUT) Oct-4/Sox2 binding sites of CDX2, followed by transfection of HEK293T cells with CDX2 overexpression plasmids, including Control vector for kidney luciferase gene (pTK-RL). Luciferase (firefly/renilla) activity was assayed 48 hours after transfection using the Dual-Glo™ Luciferase Assay System (Promega) according to the provided instructions.
ChIP assays
Cells in the lower chamber were treated with formaldehyde and sodium dodecyl sulfate. The cross-linked chromatin was isolated and treated with either a rabbit anti-CDX2 polyclonal antibody or normal rabbit immunoglobulin G overnight at 4 ℃. The deoxyribonucleic acid (DNA) was evaluated using qRT-PCR. The primers are shown in Table 1.
EMSAs
Electrophoretic mobility shift assays (EMSAs) were used to assess the regulation of Oct-4/Sox2 by CDX2. Probes with the CDX2-binding site sequence (see Table 1) were labeled with Digoxigenin-11-deoxyuridine 5-triphosphate (DIG-ddUTP). Cell protein-DNA complexes were examined with and without competitors or the anti-CDX2 antibody. After electrophoretic separation, the complexes were probed auto-radiographically using a 2nd-generation DIG Gel Shift Kit (Roche, Mannheim, Germany).
Osteoblastic induction
A StemProTM Osteogenesis Differentiation Kit (Gibco, Waltham, MA, USA) was used to measure osteoblastic induction. The cells were washed and plated in 12-well plates in mesenchymal stem cell growth medium and cultured for 5 days under standard conditions. The medium was then exchanged with complete osteogenesis differentiation medium (Invitrogen).
Alkaline phosphatase (ALP) activity
The activities of ALP in the cell lysates were determined with an ALP activity detection kit (Beyotime, Shanghai, China), and the protein levels were measured with a bicinchoninic acid kit (Cwbio, Beijing, China).
Alizarin red S staining
The cells were fixed in 4% paraformaldehyde for 30 min and stained with 1% Alizarin Red S (Cyagen, Suzhou, China) for 5 min at room temperature. Mineralized nodules were quantified after cells were incubated in 0.1 M cetylpyridinium chloride monohydrate, and calcium deposits were examined with an inverted phase contrast microscope. The absorbances were then read at 562 nm.
Adipogenic differentiation
The induction of adipogenic differentiation was undertaken using 10% fetal bovine serum (FBS), 1% Pen/strep (P/S), 0.5 mM of isobutylmethylxanthine (Sigma-Aldrich), 200 µM of indomethacin (Sigma-Aldrich), 1 µM of dexamethasone, and 10 µg/mL of insulin (Sigma-Aldrich). The cells were then grown in adipogenic maintenance medium (10 µg/mL of insulin in high-glucose Dulbecco’s modified Eagle’s medium, 10% FBS, and 1% P/S) for 3 weeks. After washing in phosphate buffered saline and fixing as describe above, the cells were stained with Oil Red O for lipid droplet detection.
Statistical analysis
All the data were analyzed using SPSS version 19.0, and the results are presented as means ± standard deviations (SDs). All the experiments were performed 3 times. Differences between the cells in the co-culture and control groups were analyzed by the Student’s t-test. The images are from 6 independent experiments. P values <0.05, <0.01, and <0.001 were considered statistically significant.
Results
Gene expression in indirectly co-cultured DPCs and PDLCs
Gene expression was determined by PCR arrays, and the DEGs between co-cultured and control cells are illustrated in the heatmaps in Figure 1A,1B. Our data indicated that the expression levels of Oct-4 and Sox2 were significantly increased in the experimental group, while the expression level of CDX2 was decreased (see Figure 1A and Table 2). Similar trends were seen in DPLCs (see Figure 1B and Table 3). We subsequently reconfirmed these changes at the protein level by western blotting (see Figure 1C-1F).
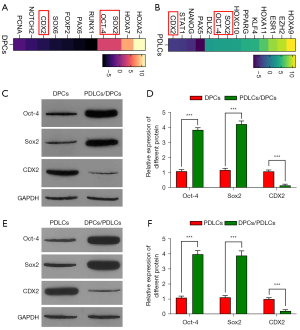
Table 2
| Gene name | Gene symbol | Fold difference (PDLCs/DPCs vs. DPCs) | P value | Functional gene grouping |
|---|---|---|---|---|
| Homeobox A2 | HOXA2 | 11.54 | 0.0328 | Segmentation/axis/symmetry |
| Embryonic development | ||||
| Homeobox A7 | HOXA7 | 7.94 | 0.0217 | Segmentation/axis/symmetry |
| Embryonic development neurogenesis | ||||
| SRY (sex determining region Y)-box 2 | SOX2 | 3.43 | 0.0240 | Somatic stem cell maintenance |
| Induced pluripotent & embryonic stem cell | ||||
| Embryonic development | ||||
| Organ morphogenesis | ||||
| Neurogenesis | ||||
| Osteogenesis | ||||
| Octamer-binding transcription factor 4 | OCT-4 | 3.25 | 0.0048 | Somatic stem cell maintenance |
| Induced pluripotent & embryonic stem cell | ||||
| Runt-related transcription factor 1 | RUNX1 | −7.94 | 0.0254 | Organ morphogenesis |
| Angiogenesis | ||||
| Hematopoiesis | ||||
| Paired box 6 | PAX6 | −7.12 | 0.0237 | Organ morphogenesis |
| Forkhead box p2 | FOXP2 | −6.46 | 0.0332 | Cell cycle |
| DNA replication | ||||
| SRY (sex determining region Y)-box 6 | SOX6 | −6.05 | 0.0043 | Hematopoiesis |
| Proliferation | ||||
| Caudal type homeobox transcription factor 2 | CDX2 | −5.16 | 0.0158 | Somatic stem cell maintenance |
| Organ morphogenesis | ||||
| Placenta development | ||||
| Axis/symmetry/segmentation | ||||
| Angiogenesis | ||||
| Notch 2 | NOTCH2 | −4.41 | 0.0442 | Segmentation/axis/symmetry |
| Embryonic development | ||||
| Organ morphogenesis | ||||
| Proliferating cell nuclear antigen | PCNA | −3.06 | 0.0132 | Other |
DPCs, dental pulp cells; PDLCs, periodontal ligament cells.
Table 3
| Gene name | Gene symbol | Fold difference (DPCs/PDLCs vs. PDLCs) | P value | Functional gene grouping |
|---|---|---|---|---|
| Homeobox A9 | HOXA9 | 11.32 | 0.0051 | Segmentation/axis/symmetry |
| Embryonic development | ||||
| Enhancer of zeste homolog 2 | EZH2 | 9.45 | 0.0096 | Cell growth |
| Resistance to apoptosis | ||||
| Estrogen receptor 1 | ESR1 | 8.48 | 0.00357 | Oncogenesis |
| Homeobox A11 | HOXA11 | 6.27 | 0.0011 | Embryonic development |
| Kruppel-like factor 4 (gut) | KLF4 | 4.35 | 0.0073 | Embryonic development |
| Ectoderm, endoderm & mesoderm formation & differentiation | ||||
| Organ morphogenesis | ||||
| Peroxisome proliferator-activated receptor gamma | PPARG | 4.16 | 0.0014 | Placenta development |
| Organ morphogenesis | ||||
| Neurogenesis | ||||
| Homeobox C10 | HOXC10 | 3.35 | 0.0022 | Segmentation/axis/symmetry |
| Embryonic development | ||||
| Neurogenesis | ||||
| SRY (sex determining region Y)-box 2 | SOX2 | 3.15 | 0.0402 | Somatic stem cell maintenance |
| Induced pluripotent & embryonic stem cell | ||||
| Embryonic development | ||||
| Organ morphogenesis | ||||
| Neurogenesis | ||||
| Osteogenesis | ||||
| Octamer-binding transcription factor 4 | OCT-4 | 3.04 | 0.0002 | Somatic stem cell maintenance |
| Induced pluripotent & embryonic stem cell | ||||
| Distal-less homeobox 2 | DLX2 | 2.83 | 0.0005 | Embryonic development |
| Organ morphogenesis | ||||
| Neurogenesis | ||||
| Paired box 5 | PAX5 | −7.28 | 0.0211 | Organ morphogenesis |
| Nanog homeobox | NANOG | −5.01 | 0.0129 | Somatic stem cell maintenance |
| Induced pluripotent & embryonic stem cell | ||||
| Embryonic development | ||||
| Signal transducer and activator of transcription 1 | STAT1 | −4.36 | 0.0124 | Hematopoiesis |
| Caudal type homeobox transcription factor 2 | CDX2 | −4.09 | 0.0202 | Somatic stem cell maintenance |
| Organ morphogenesis | ||||
| Placenta development | ||||
| Axis/symmetry/segmentation | ||||
| Angiogenesis |
PDLCs, periodontal ligament cells.
Effects of overexpression and knockdown of CDX2
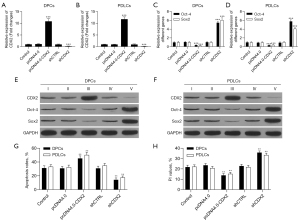
The PCR measurements showed that the CDX2 gene was effectively overexpressed and knocked down after transfection in both the DPCs and PDLCs (see Figure 2A,2B). The expression level of Sox2 and Oct-4 were significantly elevated after CDX2 knockdown and reduced on CDX2 overexpression in both cell types (see Figure 2C,2D), and similar changes were also found at the protein level (see Figure 2E,2F). CDX2 knockdown also reduced apoptosis in both DPCs and PDLCs (see Figure 2G) and reduced the ratio of cells in the synthesis stage (see Figure 2H).
CDX2 targets Oct-4/Sox2
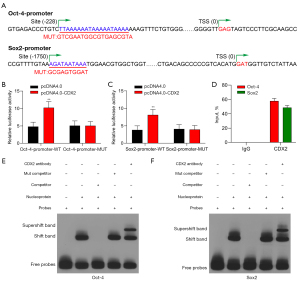
Both Oct-4 and Sox-2 were predicted by jaspar (https://jaspar.genereg.net/) to be targeted by CDX2 (see Figure 3A). The luciferase activity of Sox2 and Oct-4 mutants in 293T cells was found to be significantly lower compared to CDX2 overexpression plasmids (see Figure 3B,3C). These findings indicate that Oct-4 and Sox2 are targeted by CDX2. Chromatin Immunoprecipitation (ChIP) assays were conducted to confirm that CDX2 interacts with the Oct-4 and Sox2 promoters (see Figure 3D). As previously shown (15), the promoter levels of both genes were raised in comparison to those of the controls. Then the probes with mutations in the CDX2-Oct-4/Sox2 interaction regions were used for the EMSAs (see Figure 3E,3F). This confirmed the presence of CDX2 binding sites in both promoter regions. These interactions were eliminated by incubation with specific inhibitors, while incubation with mutant competitors had the opposite effect. Further, incubation with the anti-CDX2 antibody also reduced the interactions. These findings show that both Oct-4 and Sox2 are targeted and bound directly by CDX2.
Silencing CDX2 enhanced the odontogenic and adipogenic differentiation of DPCs
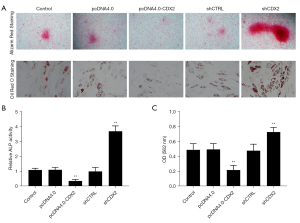
We examined odontogenic differentiation in DPCs by detecting the presence of mineralized nodules. Staining with alizarin red S showed that CDX2 knockdown significantly increased the formation of calcium deposits (see Figure 4A). Knockdown also increased the activity levels of ALP (see Figure 4B). In terms of adipogenic differentiation, CDX2 silencing led to larger numbers of lipid droplet clusters (see Figure 4A,4C).
Discussion
The use of autologous cells in regenerative treatment prevents rejection and reduces the likelihood of infection. When culturing donor cells before differentiation into the required lineage, it is critical to maximize their pluripotency. However, the ideal conditions for maintaining the stemness and pluripotency of DPCs and PDLCs in culture are not well defined. The majority of identified regulatory factors are transcription factors, such as CDX2, and cell-cycle modulators, such as Oct-4 and Sox2. In this study, we observed that CDX2 modulates osteogenesis through the direct regulation of Oct-4 and Sox2. This finding suggests new directions for the promotion of dental tissue regeneration.
CDX2 is reported to play critical roles in a variety of developmental processes. We investigated the effects of increasing the pluripotency of dental cells by co-culturing DPCs and DPLCs and observed a significant downregulation of CDX2 in the co-cultures. Mutual inhibition between CDX2 and Sox2 has been observed, and a balanced expression of the 2 genes is necessary for normal development (25). In the intestine, CDX2 modulates the proliferation-differentiation equilibrium through the regulation of the expression of genes, such as Krüppel-like factor 4, villin, mucin 2, and sucrase-isolamatase (26).
In a previous study, we demonstrated that cell-cell communication between DPCs and PDLCs enhances the differentiation potential of both cell types, as shown by increased numbers of calcified nodules, elevated ALP activity, and adipogenic differentiation (15). In the present study, we observed the significant upregulation of Oct-4 and Sox2 levels after co-cultures. These genes represent markers of pluripotency, and while their expression in differentiated cells is rare, they may be expressed in dental tissue after damage. Pluripotency may be triggered in injured dental cells by intercellular communication. A study has observed increased expression of Oct-4 and Sox2 in hypoxic PDLCs and DPCs, suggesting that hypoxia induces the expression of stem cell markers to maintain pluripotency in PDLCs and DPCs (24). Oct-4 is a POU5-family transcriptional regulator expressed in pluripotent cells where it plays a key role in the establishment of pluripotent cell populations (27) through its promotion of the expression of stem cell-associated genes. Changes in Oct-4 expression are involved in various processes of cell regeneration. Sox2 is a high-mobility group–box transcription factor, and like Oct-4, is closely involved in development (28). The results of the present study showed significant elevations of both Oct-4 and Sox2 in the co-cultured cells, indicating a potential synergistic action in the regulation of pluripotency in these cells. This suggests that odontogenic cells could potentially be used to repair dental tissue damage. However, further research is needed to determine the signaling mechanisms of the process.
Different cells have different neighbors and thus may experience different signaling triggers. A similar phenomenon has been reported in the presence of the coupling factor EphrinB2 between osteoclasts and osteoblasts, which controls osteoclast formation through interaction with EphB4 expressed in the vessel wall (29). The indirect co-cultures of DPCs and PDLCs were found to reduce the expression of CDX2 while increasing that of Sox2. Both the ChIP assays and EMSAs confirmed that CDX2 interacts with the promoter regions of the endogenous Sox2 and Oct-4, demonstrating the direct targeting of these genes by CDX2. Thus, inhibiting CDX2 expression in the co-cultured cells led to the upregulation of both Oct-4 and Sox2 and promoted the pluripotency of the cells. The function of CDX2 in epithelial differentiation is well documented, and its overexpression stimulates differentiation in intestinal epithelial cell line 6 and HT-29 cells (30), while knockdown adversely affects the polarity and morphogenesis of intestinal epithelial cells (31). Our findings verified that CDX2 reduces Oct-4 and Sox2 expression by directly binding to their promoter regions, leading to the modulation of pluripotency in dental cells. However, a study has pointed out that PDLCs have some surface antigens similar to DPCs, and the stem cell properties of PDLCs and DPCs gradually decrease with passage (32). Whether this will indirectly affect the cellular communication between PDLCs and DPCs remains to be studied.
As the influence of the culture environment is known to affect Sox2 functioning in human embryonic stem cells (33), and the activity of Sox2 is dependent on its interaction partners, it is likely that the specific actions of Sox2 are also dependent on its interaction partners. We observed that CDX2 binds to both Sox2 and Oct-4 promoters. The presence of a CDX2 binding site has also been reported to be adjacent to the Oct-4 interaction site (34), suggesting a possible cross-linkage between Oct-4 and CDX2. It is thus possible that CDX2 might de-activate Oct-4 through interactions with adjacent loci, thus reducing accessibility to Oct-4. In addition to the CDX2/Oct-4/Sox2 pathway described in this study, there are several pathways that can play a similar role. It has been reported that inhibition of LIF/STAT3 directly affects Nanog demethylation by downregulating DNA methyltransferase 1 expression (35). Other study has pointed out that STAT3 inhibits the expression of differentiation-related genes by binding to the promoter region of the Eed gene (36).
In conclusion, we showed that DPC and PDLC pluripotency is modulated by cell-cell communication. CDX2 was shown to regulate pluripotency by targeting Oct-4, while CDX2 plays an important role in modulating DPC and PDLC pluripotency by targeting Oct-4/Sox2. These results suggest a strategy for increasing pluripotency and have potential applications in the regeneration of damaged dental tissue.
Acknowledgments
Funding: The research was supported by grants from the National Natural Science Foundation of China (No. 81800954).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3492/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3492/dss
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3492/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Sun Yat-sen University (No. 2018-02-142) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Eramo S, Natali A, Pinna R, et al. Dental pulp regeneration via cell homing. Int Endod J 2018;51:405-19. [Crossref] [PubMed]
- Gruber R. Osteoimmunology: Inflammatory osteolysis and regeneration of the alveolar bone. J Clin Periodontol 2019;46:52-69. [Crossref] [PubMed]
- Ouchi T, Nakagawa T. Tissue Regeneration and Physiological Functional Recovery in Dental and Craniofacial Fields. Biomolecules 2021;11:1644. [Crossref] [PubMed]
- Hu L, Liu Y, Wang S. Stem cell-based tooth and periodontal regeneration. Oral Dis 2018;24:696-705. [Crossref] [PubMed]
- Ouchi T, Nakagawa T. Mesenchymal stem cell-based tissue regeneration therapies for periodontitis. Regen Ther 2020;14:72-8. [Crossref] [PubMed]
- Liu L, Ling J, Wei X, et al. Stem cell regulatory gene expression in human adult dental pulp and periodontal ligament cells undergoing odontogenic/osteogenic differentiation. J Endod 2009;35:1368-76. [Crossref] [PubMed]
- Ikeda H, Sumita Y, Ikeda M, et al. Engineering bone formation from human dental pulp- and periodontal ligament-derived cells. Ann Biomed Eng 2011;39:26-34. [Crossref] [PubMed]
- Lei M, Li K, Li B, et al. Mesenchymal stem cell characteristics of dental pulp and periodontal ligament stem cells after in vivo transplantation. Biomaterials 2014;35:6332-43. [Crossref] [PubMed]
- Yang C, Lee JS, Jung UW, et al. Periodontal regeneration with nano-hyroxyapatite-coated silk scaffolds in dogs. J Periodontal Implant Sci 2013;43:315-22. [Crossref] [PubMed]
- Astudillo-Ortiz E, Babo PS, Reis RL, et al. Evaluation of Injectable Hyaluronic Acid-Based Hydrogels for Endodontic Tissue Regeneration. Materials (Basel) 2021;14:7325. [Crossref] [PubMed]
- Liu Y, Gan L, Cui DX, et al. Epigenetic regulation of dental pulp stem cells and its potential in regenerative endodontics. World J Stem Cells 2021;13:1647-66. [Crossref] [PubMed]
- Coskun M. The role of CDX2 in inflammatory bowel disease. Dan Med J 2014;61:B4820. [PubMed]
- Chawengsaksophak K. Cdx2 Animal Models Reveal Developmental Origins of Cancers. Genes (Basel) 2019;10:928. [Crossref] [PubMed]
- Olsen J, Espersen ML, Jess P, et al. The clinical perspectives of CDX2 expression in colorectal cancer: a qualitative systematic review. Surg Oncol 2014;23:167-76. [Crossref] [PubMed]
- Peng Z, Liu L, Zhang W, et al. Pluripotency of Dental Pulp Cells and Periodontal Ligament Cells Was Enhanced through Cell-Cell Communication via STAT3/Oct-4/Sox2 Signaling. Stem Cells Int 2021;2021:8898506. [Crossref] [PubMed]
- Biswas A, Hutchins R. Embryonic stem cells. Stem Cells Dev 2007;16:213-22. [Crossref] [PubMed]
- Gurusamy N, Alsayari A, Rajasingh S, et al. Adult Stem Cells for Regenerative Therapy. Prog Mol Biol Transl Sci 2018;160:1-22. [Crossref] [PubMed]
- He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol 2009;25:377-406. [Crossref] [PubMed]
- Liu L, Peng Z, Huang H, et al. Luteolin and apigenin activate the Oct-4/Sox2 signal via NFATc1 in human periodontal ligament cells. Cell Biol Int 2016;40:1094-106. [Crossref] [PubMed]
- Liu L, Wei X, Ling J, et al. Expression pattern of Oct-4, Sox2, and c-Myc in the primary culture of human dental pulp derived cells. J Endod 2011;37:466-72. [Crossref] [PubMed]
- Liu L, Peng Z, Xu Z, et al. Effect of luteolin and apigenin on the expression of Oct-4, Sox2, and c-Myc in dental pulp cells with in vitro culture. Biomed Res Int 2015;2015:534952. [Crossref] [PubMed]
- Liu L, Wei X, Huang R, et al. Effect of bone morphogenetic protein-4 on the expression of Sox2, Oct-4, and c-Myc in human periodontal ligament cells during long-term culture. Stem Cells Dev 2013;22:1670-7. [Crossref] [PubMed]
- Crompton LA, McComish SF, Stathakos P, et al. Efficient and Scalable Generation of Human Ventral Midbrain Astrocytes from Human-Induced Pluripotent Stem Cells. J Vis Exp 2021; [Crossref] [PubMed]
- Zhou Y, Fan W, Xiao Y. The effect of hypoxia on the stemness and differentiation capacity of PDLC and DPC. Biomed Res Int 2014;2014:890675. [Crossref] [PubMed]
- Raghoebir L, Biermann K, Buscop-van Kempen M, et al. Disturbed balance between SOX2 and CDX2 in human vitelline duct anomalies and intestinal duplications. Virchows Arch 2013;462:515-22. [Crossref] [PubMed]
- Kazumori H, Ishihara S, Rumi MA, et al. Bile acids directly augment caudal related homeobox gene Cdx2 expression in oesophageal keratinocytes in Barrett's epithelium. Gut 2006;55:16-25. [Crossref] [PubMed]
- Bhartiya D. Are Mesenchymal Cells Indeed Pluripotent Stem Cells or Just Stromal Cells? OCT-4 and VSELs Biology Has Led to Better Understanding. Stem Cells Int 2013;2013:547501. [Crossref] [PubMed]
- Xu J, Yu L, Guo J, et al. Generation of pig induced pluripotent stem cells using an extended pluripotent stem cell culture system. Stem Cell Res Ther 2019;10:193. [Crossref] [PubMed]
- Sims NA, Walsh NC. Intercellular cross-talk among bone cells: new factors and pathways. Curr Osteoporos Rep 2012;10:109-17. [Crossref] [PubMed]
- Mallo GV, Soubeyran P, Lissitzky JC, et al. Expression of the Cdx1 and Cdx2 homeotic genes leads to reduced malignancy in colon cancer-derived cells. J Biol Chem 1998;273:14030-6. [Crossref] [PubMed]
- Gao N, Kaestner KH. Cdx2 regulates endo-lysosomal function and epithelial cell polarity. Genes Dev 2010;24:1295-305. [Crossref] [PubMed]
- Wu LP, Wei X, Ling JQ, et al. The surface antigen expression of periodontal ligament cells and dental pulp cells in vitro. Hua Xi Kou Qiang Yi Xue Za Zhi 2009;27:20-3. [PubMed]
- Puscheck EE, Awonuga AO, Yang Y, et al. Molecular biology of the stress response in the early embryo and its stem cells. Adv Exp Med Biol 2015;843:77-128. [Crossref] [PubMed]
- Erwin JA, del Rosario B, Payer B, et al. An ex vivo model for imprinting: mutually exclusive binding of Cdx2 and Oct4 as a switch for imprinted and random X-inactivation. Genetics 2012;192:857-68. [Crossref] [PubMed]
- Wang L, Jiang Z, Huang D, et al. JAK/STAT3 regulated global gene expression dynamics during late-stage reprogramming process. BMC Genomics 2018;19:183. [Crossref] [PubMed]
- Ura H, Usuda M, Kinoshita K, et al. STAT3 and Oct-3/4 control histone modification through induction of Eed in embryonic stem cells. J Biol Chem 2008;283:9713-23. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


