
Exploring the mechanism of Shexiang Tongxin dropping pill in the treatment of microvascular angina through network pharmacology and molecular docking
Introduction
Microvascular angina (MVA) is a group of clinical manifestations including angina pectoris or angina-like chest pain, positive exercise test, and exclusion of epicardial coronary artery spasm; however, coronary angiography (CAG) examination does not reveal significant stenosis of epicardial vessels (1). Epidemiological surveys have shown that MVA accounts for approximately 10–15% of angina pectoris cases; the incidence of MVA can be as high as 30% in patients with nonobstructive stable angina pectoris (2). In patients with suspected myocardial ischemia and indication for CAG, about 41% of women and 8% of men have been found to have no significant epicardial stenosis (3). Although the coronary microcirculation system predominantly consists of microarterial capillaries and microvenules with a diameter of ≤150 µm, CAG can only show vessels >500 µm in diameter. The main clinical indices used to assess coronary microcirculation are microvascular resistance reserve (MRR), resistive reserve ratio (RRR), coronary flow reserve (CFR), and resting and hyperemic mean transit time (Tmn) (4). The major drugs currently used to treat MVA are nitrates and beta-blockers. Beta-blockers (propranolol, nebivolol, and carvedilol) have been reported to improve exercise tolerance and symptoms, and are up to 75% effective in the treatment MVA (5). Although MVA is generally associated with a good prognosis, a related study showed that 20% of patients with MVA experience worsening symptoms, which greatly increases the incidence of adverse cardiovascular events (6). Coronary microcirculation disorder (CMD) is the main cause of MVA (7). Microcirculatory disorders are disorders of structure and function and mainly include microcirculatory embolism and endothelial cell dysfunction (8). Structural or functional abnormalities of the CMD, resulting in impaired vasodilation, as evidenced by the inability of the coronary arteries to expand appropriately to accommodate myocardial oxygen demand (9). This might explain the symptoms of ischemia in MVA patients without significant epicardial stenosis.
In recent years, the concept of multitargeted and comprehensive treatment of diseases with traditional Chinese medicine (TCM) has witnessed great progress. In the cardiovascular field, the importance of TCM is increasing. It is not uncommon for patients to experience adverse reactions to the simple Western medicine intervention, and the curative effect is limited. Unlike Western medicine, TCM comprises a multitargeted, multi-pathway, and multilinked treatment model. The main ingredients of the Shexiang Tongxin dropping pill (STDP) are musk (she xiang), radix salviae (dan shen), borneol (bing pian), toad skin secretion cake (chan su), calculus bovis (niu huang), folium ginseng (ren shen ye), and bear gall (xiong dan) (10). It has been demonstrated that STDP can alleviate CMDs, likely via anti-inflammatory, antioxidant, and apoptosis-inhibiting mechanisms (11). The composition of STDP is complex, and due to various constraints, its mechanism of action has not been fully elucidated. We employed network pharmacology and molecular docking techniques to investigate the action mechanism of STDP in the treatment of MVA.
TCMSP is a unique systems pharmacology platform of Chinese herbal medicines that captures the relationships between drugs, targets and diseases (https://tcmsp-e.com/tcmsp.php). HERB (BenCaoZuJian as its Chinese name), a high-throughput experiment- and reference-guided database of TCM (http://herb.ac.cn). We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3976/rc).
Methods
Schematic diagram
Figure 1 presents the schematic diagram of the study design and workflow. We employed network pharmacology and molecular docking to investigate the action mechanism of STDP against MVA.
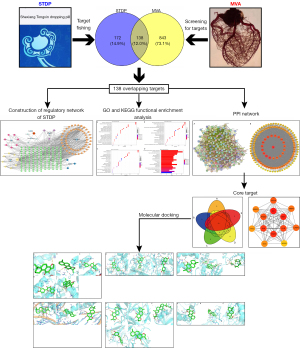
Screening the main active ingredients and targets of STDP
The compounds of borneol, radix salviae, musk, toad skin secretion cake, calculus bovis, folium ginseng, and bear gall in STDP were collected from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) and HERB databases (12,13). In addition, we searched for relevant literature through PubMed to supplement the drug composition and target of action (14-16). Oral bioavailability (OB) and drug-likeness (DL) are essential indicators targeted to determine the absorption, distribution, metabolism, and excretion (ADME) of drug metabolism kinetics (17). The desired compounds were screened for active ingredients with an OB ≥30% and DL ≥0.18, and the targets of the corresponding compounds’ actions were obtained through the TCMSP database. In addition, the compounds that met the screening criteria but did not possess the corresponding target in the TCMSP database were selected and their canonical simplified molecular-input line-entry system (SMILES) numbers were screened from the PubChem database with reference to the Chemical Abstracts Service (CAS) numbers of these compounds. “Homo sapiens” was selected, and the top 15 predicted targets with probability >0 were added by SwissTargetPrediction (18). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
MVA disease target collection
The search entry for the target disease was “Microvascular Angina”, and its Medical Subject Headings (MeSH) Unique ID (D017566) was obtained from the MeSH database in PubMed. We searched the GeneCards database (19), Online Mendelian Inheritance in Man (OMIM) database (20), Pharmacogenetics and Pharmacogenomics Knowledge Base (PharmGKB) database (21), DisGeNET (22), and Therapeutic Target Database (TTD) (23) using “Microvascular Angina” as the keyword. Duplicate targets were deleted and the target data for the disease was created.
Acquisition of potential targets for STDP against MVA
The software Rstudio 4.1.6 (RStudio, Boston, MA, USA) and the “ggvenn” package were used to draw the Venn diagram. The targets of action of the active ingredients of STDP were compared with the targets of MVA to obtain the intersection targets. These intersection targets served as the potential targets of STDP for MVA treatment.
STDP active ingredient-MVA-target interaction network construction
We removed the active ingredients from STDP that did not intersect with the disease target. All remaining active ingredients and the intersecting targets were then imported into Cytoscape 3.9.1 (24). The topology of the STDP-regulated MVA network was structured in accordance with the drug-compound-target partition. The node size was set to be continuously mapped by the degree of connectivity. Each node represented a drug, compound, or target. Connections between the nodes represented the presence of interactions between them. The degree of the node represented the number of nodes connected.
Enrichment analyses
The software Rstudio 4.1.3 and the “org.Hs.eg.db” package of the Bioconductor database was used to convert the gene symbols of the intersection targets into Entrez IDs. We then used the “ClusterProfiler” package to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the network (25). We set P<0.01 and false discovery rate (FDR) <0.01 as the entries to plot the results of GO and KEGG enrichment analysis, separately. We also identified the top 20 cellular components (CC), biological processes (BP), molecular functions (MF), and signaling pathways involved in the intersecting targets.
Construction of the protein-protein interaction (PPI) network
The PPI network analysis of the intersecting targets was performed by the Search Tool for the Retrieval of Interacting Genes/Genomes (STRING) database, and “Homo sapiens”, which showed the lowest interaction score (≥0.4), was used as the screening criterion (26). The data was exported in the TSV format and imported into Cytoscape 3.9.1 for further analyses.
The core target screen
The PPI network was screened using the cytoHubba plugin (27). We then selected the top 20 core genes by maximal clique centrality (MCC), degree, edge-percolated component (EPC), closeness, and radiality. After selecting the top 20 core genes, the intersection was performed to obtain the key targets, which were then arranged in the descending order by MCC owing to the superior confidence of MCC in cytoHubba.
Molecular docking
We downloaded the 3-dimensional (3D) structures of core targets from the Protein Data Bank (PDB) (28). Next, we obtained the SDF structure files of the compounds through the PubChem database (29). We also used Open Babel 3.1.1 to convert the SDF file into a PDB file (30). Further, PYMOL 2.4.0 (31) was used to dehydrate the receptor protein, remove the ligand, and retain the single chain, and the AutoDock Tools (32) were used to modify the receptor protein by hydrogenation and charge balancing.
Statistical analyses
The data were summarized using Windows, WPS Office version (11.1.0.11744)-Release; the PPI data were analyzed by Cytoscape 3.9.1; the protein processing data were molecularly docked using the AutoDock Vina 1.1.2. The pathway enrichment visualization analysis was performed from Rstudio 4.1.6. All relevant data analysis results are presented in the “Results” section.
Results
Screening the active ingredients and targets of STDP
The pinyin herbal names bing pian, dan shen, niu huang, and ren shen ye were entered in the TCMSP database, and chan su and xiong dan were entered into the HERB database, with the parameters set to OB ≥30% and DL ≥0.18. The active ingredients of she xiang were supplemented by reviewing the relevant literature in recent years. The SwissTargetPrediction database was used to predict the target points of she xiang, and those active ingredients that failed to match the target in the TCMSP database and the setting of “high” gastrointestinal absorption were chosen. The targets of action of some of the active ingredients were supplemented by searching the relevant literature in PubMed. In all, 93 active ingredients were obtained, including 7 for she xiang, 9 for ren shen ye, 65 for dan shen, 5 for niu huang, 4 for chan su, 3 for bing pian, and 1 for xiong dan. Among them, deoxycholic acid is a common component of xiong dan and niu huang. A total of 310 target genes corresponded to the active ingredients of STDP after removing the duplicates.
Acquisition of potential targets for STDP against MVA
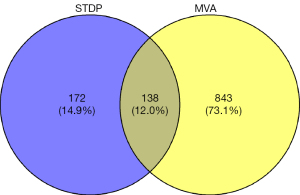
A total of 1,024 MVA disease targets were obtained by searching GeneCards (score ≥1), TTD, DisGeNET, OMIM, and PharmGKB databases, and 981 remained after removal of duplicates. The active ingredient of STDP regulates targets that intersect with MVA disease genes are the potential targets of STDP against MVA. The targets corresponding to the active ingredients of the drugs were intersected with the disease targets. A total of 138 intersecting targets were obtained, which constituted the potential targets of STDP for MVA treatment (Figure 2). After the removal of duplicates, the total number of active ingredients corresponding to these targets was 81.
STDP active ingredient-MVA-target interaction network construction
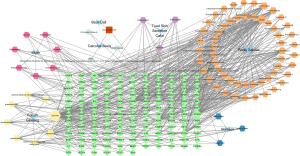
An STDP active ingredient-MVA-target interaction network was constructed based on 138 target genes and their active ingredients using Cytoscape 3.9.1 (Figure 3). The network consisted of 226 nodes and 607 edges. The node size was proportional to the degree value. The blue concave quadrilateral contained 7 of the main components of STDP, namely musk (she xiang), toad skin secretion cake (chan su), radix salviae (dan shen), calculus bovis (niu huang), borneol (bing pian), folium ginseng (ren shen ye), and bear gall (xiong dan). The red square (one only) was a component of deoxycholic acid, the common component of both bear gall and calculus bovis. The positive hexagon represented the compound, and yellow, pink, cyan, purple, orange, and dark blue were the active ingredients of folium ginseng, musk, calculus bovis, toad skin secretion cake, radix salviae, and borneol, respectively. Green diamonds were the intersection targets of STDP and MVA.
Enrichment analyses
The GO and KEGG pathway enrichment analysis of the 138 intersectional targets was performed using RStudio 4.1.3. Furthermore, P<0.01 and FDR <0.01 were set for the entries to plot the results of GO and KEGG enrichment analyses separately. The top 20 CC, BP, MF, and signaling pathways involved in the intersecting targets were identified (Figure 4).
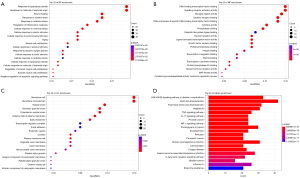
The GO-BP of the targets included a response to lipopolysaccharides, response to molecules of bacterial origin, wound healing, response to oxidative stress, aging, muscle cell proliferation, and negative regulation of the apoptotic signaling pathway (Figure 4A).
The GO-MF comprised signaling receptor activator activity, receptor-ligand activity, nuclear receptor activity, growth factor receptor binding, protein phosphatase binding, and mitogen-activated protein (MAP) kinase activity (Figure 4B).
The GO-CC consisted of membrane raft, membrane microdomain, vesicle lumen, external side of the plasma membrane, focal adhesion, organelle outer membrane, and platelet alpha granule lumen (Figure 4C).
The potential target genes of STDP in MVA were predominantly enriched in the advanced glycation end products/receptor for advanced glycation end products (AGE-RAGE) signaling pathway, lipids and atherosclerosis, fluid shear stress and atherosclerosis, tumor necrosis factor (TNF) signaling pathway, interleukin (IL)-17 signaling pathway, hypoxia-inducible factor (HIF)-1 signaling pathway, and C-type lectin receptor signaling pathway (Figure 4D). Each pathway consisted of different targets (Table 1).
Table 1
| ID | Pathway | Gene |
|---|---|---|
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | AKT1, BAX, CCND1, BCL2, CASP3, MAPK14, EDN1, F3, ICAM1, IL1A, IL1B, IL6, CXCL8, JUN, MMP2, NFKB1, NOS3, SERPINE1, PRKCA, PRKCB, MAPK1, MAPK3, MAPK8, MAPK9, RELA, CCL2, SELE, STAT1, STAT3, THBD, TNF, VCAM1, VEGFA, CAV1, GSTM1, GSTP1, HMOX1, HSP90AA1, IFNG, ITGB3, MMP9, NFE2L2, PLAT, TP53, NCF1 |
| hsa05417 | Lipid and atherosclerosis | AKT1, BAX, BCL2, BCL2L1, CASP1, CASP3, CASP7, CASP8, CASP9, CD40LG, MAPK14, HSP90AA1, ICAM1, IL1B, IL6, CXCL8, JUN, LDLR, MMP1, MMP3, MMP9, MYD88, NFE2L2, NFKB1, NFKBIA, NOS3, PPARG, PRKCA, MAPK1, MAPK3, MAPK8, MAPK9, RELA, CCL2, SELE, STAT3, TLR4, TNF, TP53, VCAM1, VLDLR, NCF1, BIRC5, CDKN1A, PCNA, PRKCB, RAF1, RB1, STAT1, EDN1, CXCL10, IRF1, PTGS2, IFNG, IL4, IL10, NOS2, IL2, SERPINE1, IL1A, PTPN6, CTSD, VDR, MDM2, CXCL11, SPP1, EGFR, P2RX7, ROCK1, MMP2, VEGFA, AHR, HIF1A, EGF, ACP1, GSTM1, HMOX1, MET, PTPN1, INSR, PPARA, SREBF1, ITGB3, CCND1, ESR1, ESR2, APP, MAPT, MME, CXCR3, ADRB2, EDNRA, HTR1B, PDE4D, MPO, PPARD, CAV1 |
| hsa05418 | Fluid shear stress and atherosclerosis | AKT1, BCL2, CAV1, MAPK14, EDN1, GSTM1, GSTP1, HMOX1, HSP90AA1, ICAM1, IFNG, IL1A, IL1B, ITGB3, JUN, MMP2, MMP9, NFE2L2, NFKB1, NOS3, PLAT, MAPK8, MAPK9, RELA, CCL2, SELE, THBD, TNF, TP53, VCAM1, VEGFA, NCF1 |
| hsa05161 | Hepatitis B | AKT1, BIRC5, BAX, BCL2, CASP3, CASP8, CASP9, CDKN1A, MAPK14, IL6, CXCL8, JUN, MMP9, MYD88, NFKB1, NFKBIA, PCNA, PRKCA, PRKCB, MAPK1, MAPK3, MAPK8, MAPK9, RAF1, RB1, RELA, STAT1, STAT3, TLR4, TNF, TP53 |
| hsa04668 | TNF signaling pathway | AKT1, CASP3, CASP7, CASP8, MAPK14, EDN1, ICAM1, IL1B, IL6, CXCL10, IRF1, JUN, MMP3, MMP9, NFKB1, NFKBIA, MAPK1, MAPK3, MAPK8, MAPK9, PTGS2, RELA, CCL2, SELE, TNF, VCAM1 |
| hsa04657 | IL-17 signaling pathway | CASP3, CASP8, MAPK14, HSP90AA1, IFNG, IL1B, IL4, IL6, CXCL8, CXCL10, JUN, MMP1, MMP3, MMP9, NFKB1, NFKBIA, MAPK1, MAPK3, MAPK8, MAPK9, PTGS2, RELA, CCL2, TNF |
| hsa05215 | Prostate cancer | AKT1, AR, CCND1, BCL2, CASP9, CDKN1A, EGF, EGFR, ERBB2, GSTP1, HSP90AA1, MDM2, MMP3, MMP9, NFKB1, NFKBIA, PLAT, PLAU, MAPK1, MAPK3, RAF1, RB1, RELA, TP53 |
| hsa04066 | HIF-1 signaling pathway | AKT1, BCL2, CDKN1A, EDN1, EGF, EGFR, ERBB2, HIF1A, HMOX1, IFNG, IL6, INSR, NFKB1, NOS2, NOS3, SERPINE1, PRKCA, PRKCB, MAPK1, MAPK3, RELA, STAT3, TLR4, VEGFA |
| hsa05205 | Proteoglycans in cancer | AKT1, CCND1, CASP3, CAV1, CDKN1A, MAPK14, EGFR, ERBB2, ESR1, FGF2, HIF1A, IGF2, ITGB3, MDM2, MET, MMP2, MMP9, PLAU, PRKCA, PRKCB, MAPK1, MAPK3, PTPN6, RAF1, ROCK1, STAT3, TLR4, TNF, TP53, VEGFA, AR, BCL2, CASP9, EGF, GSTP1, HSP90AA1, MMP3, NFKB1, NFKBIA, PLAT, RB1, RELA, EDN1, HMOX1, IFNG, IL6, INSR, NOS2, NOS3, SERPINE1, ADRB2, AHR, BIRC5, ESR2, GSTM1, JUN, PPARA, VDR, BCL2L1, FGF1, IL2, IL4, SPP1, HSPB1, IL1A, IL1B, MAPT, MYD88, MAPK8, MAPK9, BAX, CXCL8, MMP1, PTGS2, NFE2L2, STAT1, CNR1, CTSD, LDLR |
| hsa05145 | Toxoplasmosis | AKT1, BCL2, BCL2L1, CASP3, CASP8, CASP9, CD40LG, MAPK14, IFNG, IL10, LDLR, MYD88, NFKB1, NFKBIA, NOS2, MAPK1, MAPK3, MAPK8, MAPK9, RELA, STAT1, STAT3, TLR4, TNF |
| hsa05133 | Pertussis | CASP1, CASP3, CASP7, MAPK14, IL1A, IL1B, IL6, CXCL8, IL10, IRF1, JUN, MYD88, NFKB1, NOS2, MAPK1, MAPK3, MAPK8, MAPK9, RELA, TLR4, TNF |
| hsa05212 | Pancreatic cancer | AKT1, BAX, CCND1, BCL2L1, CASP9, CDKN1A, EGF, EGFR, ERBB2, NFKB1, MAPK1, MAPK3, MAPK8, MAPK9, RAF1, RB1, RELA, STAT1, STAT3, TP53, VEGFA |
| hsa05163 | Human cytomegalovirus infection | AKT1, BAX, CCND1, CASP3, CASP8, CASP9, CDKN1A, MAPK14, EGFR, IL1B, IL6, CXCL8, ITGB3, MDM2, NFKB1, NFKBIA, PRKCA, PRKCB, MAPK1, MAPK3, PTGER4, PTGS2, RAF1, RB1, RELA, ROCK1, CCL2, STAT3, TNF, TP53, VEGFA, AR, BCL2, EGF, ERBB2, GSTP1, HSP90AA1, MMP3, MMP9, PLAT, PLAU, BCL2L1, MAPK8, MAPK9, STAT1, IFNG, CXCL10, LDLR, PPARA, ESR1, ESR2, JUN, MMP2, MMP1, IL2, IL4, MYD88, MET, BIRC5, AHR, NFE2L2, IRF1, SPP1, GSTM1, HMOX1, IGF2, FGF1, FGF2, NOS2, ICAM1, CHEK2, INSR |
| hsa05140 | Leishmaniasis | MAPK14, IFNG, IL1A, IL1B, IL4, IL10, JUN, MYD88, NFKB1, NFKBIA, NOS2, PRKCB, MAPK1, MAPK3, PTGS2, PTPN6, RELA, STAT1, TLR4, TNF, NCF1 |
| hsa05142 | Chagas disease | AKT1, CASP8, MAPK14, IFNG, IL1B, IL2, IL6, CXCL8, IL10, JUN, MYD88, NFKB1, NFKBIA, NOS2, SERPINE1, MAPK1, MAPK3, MAPK8, MAPK9, RELA, CCL2, TLR4, TNF |
| hsa05167 | Kaposi sarcoma-associated herpesvirus infection | AKT1, BAX, CCND1, CASP3, CASP8, CASP9, CDKN1A, MAPK14, FGF2, HIF1A, ICAM1, IL6, CXCL8, JUN, NFKB1, NFKBIA, MAPK1, MAPK3, MAPK8, MAPK9, PTGS2, RAF1, RB1, RELA, STAT1, STAT3, TP53, VEGFA |
| hsa04625 | C-type lectin receptor signaling pathway | AKT1, CASP1, CASP8, MAPK14, IL1B, IL2, IL6, IL10, IRF1, JUN, MDM2, NFKB1, NFKBIA, MAPK1, MAPK3, MAPK8, MAPK9, PTGS2, RAF1, RELA, STAT1, TNF |
| hsa05219 | Bladder cancer | CCND1, CDKN1A, EGF, EGFR, ERBB2, CXCL8, MDM2, MMP1, MMP2, MMP9, MAPK1, MAPK3, RAF1, RB1, TP53, VEGFA |
| hsa05164 | Influenza A | AKT1, BAX, CASP1, CASP3, CASP8, CASP9, ICAM1, IFNG, IL1A, IL1B, IL6, CXCL8, CXCL10, MYD88, NFKB1, NFKBIA, PRKCA, PRKCB, MAPK1, MAPK3, RAF1, RELA, CCL2, STAT1, TLR4, TNF |
| hsa01522 | Endocrine resistance | AKT1, BAX, CCND1, BCL2, CDKN1A, MAPK14, EGFR, ERBB2, ESR1, ESR2, JUN, MDM2, MMP2, MMP9, MAPK1, MAPK3, MAPK8, MAPK9, RAF1, RB1, TP53 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; AGE-RAGE, advanced glycation end products/receptor for advanced glycation end products.
Construction of the PPI network
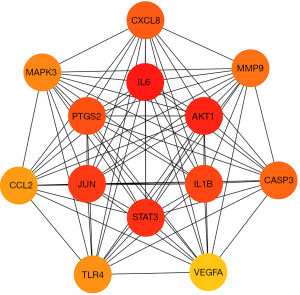
A Venn diagram was plotted to obtain the STDP and MVA intersection targets using RStudio 4.1.3 and the “ggvenn” package. A total of 138 common targets were obtained as outcomes (Figure 2). A PPI network analysis of the intersecting targets was performed using the STRING (https://cn.string-db.org/) database, and the species “Homo sapiens” and the lowest interaction score of ≥0.4 were used as the screening criteria. The PPI network diagram generated by the STRING database was saved, with each edge representing a PPI (Figure 5A). The PPI network information generated from the STRING database was exported in TSV format and imported into Cytoscape 3.9.1 to further construct the network for subsequent core target screening (Figure 5B). The higher the number of lines in the network (Figure 5A,5B), the greater the correlation and the higher the ranking of the target in the PPI network. The network (Figure 5B) contained 138 nodes and 2,966 edges, and their distribution sizes were arranged according to degree values. The top 20 core genes were selected by “MCC”, “Degree”, “EPC”, “Closeness”, and “Radiality” (Table 2). The intersecting targets were then taken to further filter the key targets by plotting the Venn diagram using RStudio 4.1.6 and the “VennDiagram” package (Figure 6). The top 20 genes were screened using cytoHubba (MCC, Degree, EPC, Closeness, and Radiality), following which the intersection was taken. A total of 13 key targets were finally obtained (Figure 6), which were ranked sequentially in MCC descending order (Table 3), as follows: IL6, AKT1, STAT3, JUN, IL-1β, PTGS2, CXCL8, CASP3, MMP9, MAPK3, TLR4, CCL2, and VEGFA. Extraction of the above 13 key targets from the PPI network created a subset to further construct the PPI core network (Figure 7), which contained 13 nodes and 78 edges. The network followed the MCC descending order from red to yellow gradient, with red indicating a higher ranking in the MCC. The top rank was for IL-6, whereas the bottom rank was for VEGFA.
Table 2
| Rank | MCC | Degree | EPC | Closeness | Radiality |
|---|---|---|---|---|---|
| 1 | TNF | CCL2 | AKT1 | TNF | TNF |
| 2 | IL6 | IL6 | VEGFA | IL6 | IL6 |
| 3 | AKT1 | AKT1 | CASP3 | AKT1 | AKT1 |
| 4 | STAT3 | IL1β | IL1β | IL1β | IL1β |
| 5 | JUN | VEGFA | TNF | VEGFA | VEGFA |
| 6 | IL1β | TP53 | IL6 | TP53 | TP53 |
| 7 | PTGS2 | MAPK3 | MMP9 | MAPK3 | MAPK3 |
| 8 | CXCL8 | STAT3 | STAT3 | STAT3 | STAT3 |
| 9 | CASP3 | JUN | EGFR | JUN | JUN |
| 10 | MMP9 | EGFR | MAPK3 | EGFR | EGFR |
| 11 | IL10 | CASP3 | JUN | CASP3 | CASP3 |
| 12 | MAPK3 | MMP9 | TP53 | MMP9 | PPARG |
| 13 | TLR4 | PPARG | PTGS2 | PPARG | MMP9 |
| 14 | CCL2 | PTGS2 | EGF | PTGS2 | PTGS2 |
| 15 | IL4 | EGF | TLR4 | EGF | EGF |
| 16 | IFNG | HIF1A | PPARG | HIF1A | HIF1A |
| 17 | ICAM1 | CCL2 | CXCL8 | CCL2 | CCL2 |
| 18 | VEGFA | CXCL8 | CCL2 | CXCL8 | ESR1 |
| 19 | VCAM1 | ESR1 | HIF1A | ESR1 | CXCL8 |
| 20 | IL1A | TLR4 | IL10 | TLR4 | TLR4 |
MCC, maximal clique centrality; EPC, edge-percolated component.
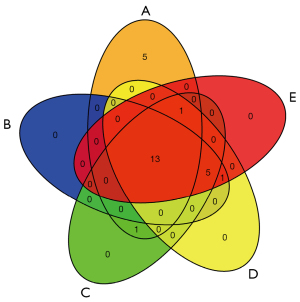
Table 3
| Entrez ID | Target | Description |
|---|---|---|
| 3569 | IL-6 | Interleukin-6 |
| 207 | AKT1 | RAC-alpha serine/threonine-protein kinase |
| 6774 | STAT3 | Signal transducer and activator of transcription 3 |
| 3725 | JUN | Transcription factor Jun |
| 3553 | IL-1β | Interleukin-1 beta |
| 5743 | PTGS2 | Prostaglandin G/H synthase 2 |
| 3576 | CXCL8 | Interleukin-8 |
| 836 | CASP3 | Caspase-3 |
| 4318 | MMP9 | Matrix metalloproteinase-9 |
| 5595 | MAPK3 | Mitogen-activated protein kinase 3 |
| 7099 | TLR4 | Toll-like receptor 4 |
| 6347 | CCL2 | C-C motif chemokine 2 |
| 7422 | VEGFA | Vascular endothelial growth factor A |
IL, interleukin; AKT1, RAC-alpha serine/threonine-protein kinase; STAT3, signal transducer and activator of transcription 3; JUN, transcription factor Jun; PTGS2, prostaglandin G/H synthase 2; CXCL8, interleukin-8; CASP3, caspase-3; MMP9, matrix metalloproteinase-9; MAPK3, mitogen-activated protein kinase 3; TLR4, Toll-like receptor 4; CCL2, C-C motif chemokine 2; VEGFA, vascular endothelial growth factor A.
Molecular docking
The top 5 targets in the PPI core network were subjected to molecular docking. In addition, our previous study showed that TLR4 was closely associated with CMD (33); hence, molecular docking of TLR4 was performed in further studies. The more stable the binding conformation, the lower the binding energy required, with a binding energy of ≤−4.25 kcal/mol implying weak binding activity, ≤−5.0 kcal/mol indicating excellent binding activity, and ≤−7.0 kcal/mol indicating strong binding activity (34). Molecular docking of IL6, AKT1, STAT3, JUN, IL-1β, and TLR4 and their corresponding compounds was performed, and the results are presented in Table 4. Subsequently, 3D docking schematics of the docking results were drawn using PYMOL 2.4.0.
Table 4
| Target | Entry | PDB ID | Molecular name | Docking score (kcal/mol) |
|---|---|---|---|---|
| IL-6 | P05231 | 1ALU | Luteolin | −7.1 |
| Dihydrotanshinone I | −7.1 | |||
| Cryptotanshinone | −7.2 | |||
| Tanshinone IIA | −7.5 | |||
| Quercetin | −7.1 | |||
| AKT1 | P31749 | 7NH5 | Luteolin | −10 |
| Kaempferol | −9.8 | |||
| Quercetin | −9.3 | |||
| STAT3 | P40763 | 6TLC | Cryptotanshinone | −7.8 |
| Ginsenoside-F2 | −6.8 | |||
| Ginsenoside Rg7 | −7.8 | |||
| JUN | P05412 | 1JUM | Dihydrotanshinone I | −8.7 |
| Cryptotanshinone | −8.6 | |||
| Tanshinone IIA | −8.2 | |||
| IL-1β | P01584 | 6Y8M | Dihydrotanshinone I | −7.5 |
| Cryptotanshinone | −7.7 | |||
| Tanshinone IIA | −7.7 | |||
| Ginsenoside Rh2 | −7.4 | |||
| Quercetin | −6.6 | |||
| TLR4 | O00206 | 2Z62 | Dihydrotanshinone I | −6.9 |
| Cryptotanshinone | −7 | |||
| Tanshinone IIA | −6.8 |
IL, interleukin; AKT1, RAC-alpha serine/threonine-protein kinase; STAT3, signal transducer and activator of transcription 3; JUN, transcription factor Jun; TLR4, Toll-like receptor 4.
Interaction analysis of the active ingredients with the IL6 receptor
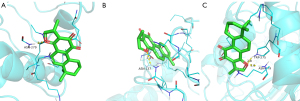
The molecular docking results of luteolin, dihydrotanshinone I, cryptotanshinone, tanshinone IIA, and quercetin with the IL6 receptor revealed a strong binding ability of these five compounds to the IL6 receptor. The nucleophilic capacity was tanshinone IIA > cryptotanshinone > luteolin, dihydrotanshinone I, and quercetin. The potential binding sites were mainly LYS-66, GLN-156, ASP-34, GLN-175, ARG-179, ARG-30, ASN-63, and TYR-97. All five compounds were able to form at least one hydrogen bond with the amino acids, and the hydrogen bond distances were relatively short, averaging 2.4 Å (Figure 8). Particularly, luteolin formed four hydrogen bonds with the active residues of the IL6 receptor, which is greater than those formed by the other four compounds, thereby indicating that this compound binds more strongly to the protein binding pocket (Figure 8C).
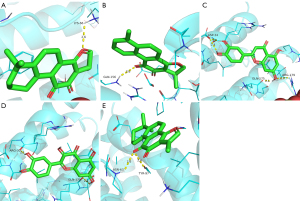
Interaction analysis of the active ingredients with the AKT1 receptor
The docking results of luteolin, kaempferol, and quercetin with AKT1 indicated the strong binding ability of the three compounds with the receptor. The nucleophilic capacity was luteolin > kaempferol > quercetin. The potential binding sites were chiefly ASN-53, ASN-204, SER-205, THR-291, and THR-211. All three compounds formed at least one hydrogen bond with the amino acids, and the hydrogen bond distances were relatively short, averaging 2.4 Å (Figure 9). Additionally, the results showed that ASN-204, SER-205, and THR-291 were the common sites for kaempferol and luteolin, which alluded that these two compounds exhibit high similarities and similar binding energies (Figure 9B,9C).
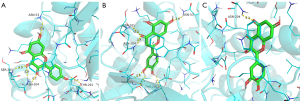
Interaction analysis of the active ingredients with the STAT3 receptor
The docking results of cryptotanshinone, ginsenoside Rg7, and ginsenoside-F2 with the STAT3 receptor demonstrated the strong binding ability of the three compounds with the receptor. The nucleophilic capacity was cryptotanshinone, ginsenoside Rg7 > ginsenoside-F2. The potential binding sites were mainly ARG-215, ARG-325, GLY-253, ASP-334, PRO-333, GLU-407, LYS-409, SER-2, SER-85, TYR-82, and GLY-295. All three compounds formed multiple hydrogen bonds with the amino acids, and the hydrogen bond distances were short, averaging 2.4 Å (Figure 10). The results signified that ginsenoside Rg7 formed eight hydrogen bonds with the active residues of the STAT3 receptor, which is higher than those formed by the other two compounds, thereby suggesting that this compound binds more strongly to the protein pocket (Figure 10B).
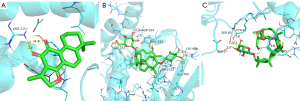
Interaction analysis of the active ingredients with the JUN receptor
The docking results of cryptotanshinone, dihydrotanshinone I, and tanshinone IIA with the JUN receptor indicated the strong binding ability of the three compounds with the receptor. The nucleophilic capacity was dihydrotanshinone I > cryptotanshinone > tanshinone IIA. The potential binding sites were mainly DC-210, DC-310, ARG-270, DA-309, and DG-208. DC-310 was the common binding site of these three compounds to the JUN receptor, thus implying a high similarity among them. All three compounds formed multiple hydrogen bonds with the amino acids, and the bond distances were short, averaging 2.3 Å (Figure 11). The results showed that dihydrotanshinone I formed five hydrogen bonds with the active residues of the JUN receptor, which is higher than those formed by the other two compounds, suggesting that this compound binds more strongly to the protein pocket (Figure 11B).
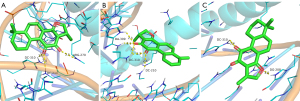
Interaction analysis of the active ingredients with the IL-1β receptor
The docking results of cryptotanshinone, dihydrotanshinone I, ginsenoside Rh2, quercetin, and tanshinone IIA with the IL-1β receptor revealed the strong binding ability of the five compounds with the receptor. The nucleophilic capacity was cryptotanshinone, tanshinone IIA > dihydrotanshinone I > ginsenoside Rh2 > quercetin. The potential binding sites were chiefly LYS-77, SER-125, LYS-77, ARG-11, GLN-39, GLU-37, GLU-4, ASN-7, and TYR-68. All five compounds formed at least one hydrogen bond with the amino acids, and the bond distances were relatively short, averaging 2.3 Å (Figure 12). The findings indicated that ginsenoside Rh2 formed five hydrogen bonds with the active residues of the IL-1β receptor, which is greater than those formed by the other two compounds, thereby signifying that this compound binds more strongly to the protein pocket (Figure 12C).
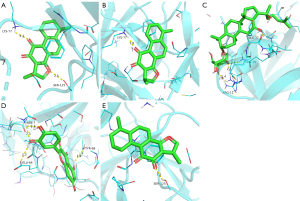
Interaction analysis of the active ingredients with the TLR4 receptor
The docking results of cryptotanshinone, dihydrotanshinone I, and tanshinone IIA with the TLR4 receptor established the strong binding ability of the three compounds with the receptor. The nucleophilic capacity was cryptotanshinone > dihydrotanshinone I > tanshinone IIA. The potential binding sites were predominantly ASN-279, ASN-137, and TRP-275. ASN-279 was a common site for cryptotanshinone and dihydrotanshinone I, thus indicating the strong structural similarity between the two compounds (Figure 13A,13B). All three compounds formed at least one hydrogen bond with the amino acids, and the bond distances were relatively short, averaging 2.3 Å (Figure 13).
Discussion
Presently, CMD, which is a structural or functional abnormality of the coronary microcirculatory system, is considered the main mechanism resulting in MVA (35). Endothelial dysfunction, microcirculatory embolism, smooth muscle dysfunction, and autonomic dysfunction are the main mechanisms leading to microcirculatory disorders (36). These factors contribute to the occurrence of MVA as a result of each other. In this study, the potential mechanism of action of STDP against MVA was analyzed using modern bioinformatics tools. It was revealed that STDP possibly exerts its therapeutic effects via processes such as anti-inflammation, immunomodulation, lipid metabolism, promotion of smooth muscle cell proliferation and differentiation, and regulation of cellular autophagy.
In this study, the regulatory network of STDP against MVA was constructed, and the results showed that STDP treatment of MVA was multicomponent, multitargeted, and multilinked. Subsequently, GO and KEGG enrichment analyses were performed on the 138 targets at the intersection of STDP and MVA. The results of GO enrichment analysis suggested that STDP could be engaged in the regulation of response to lipopolysaccharides, response to molecules of bacterial origin, wound healing, response to oxidative stress, aging, muscle cell proliferation, and negative regulation of apoptotic signaling pathway against MVA. Additionally, STDP could affect the expression of genes by interfering with signaling receptor activator activity, receptor-ligand activity, nuclear receptor activity, growth factor receptor binding, protein phosphatase binding, and MAP kinase activity. Largely, STDP prevents MVA by regulating the AGE-RAGE signaling pathway in diabetic complications, lipids and atherosclerosis, fluid shear stress and atherosclerosis, the TNF signaling pathway, the IL-17 signaling pathway, the HIF-1 signaling pathway, and the C-type lectin receptor signaling pathway. Notably, RAGE has an important biological role as a member of the innate immune system (37). As the AGE-RAGE signaling pathway is activated, it affects the cellular response to various stimuli and increases cell damage. These comprise a variety of pro-inflammatory pro-atherogenic mediators, including nuclear factor-kappaB (NF-κB) dependent mediators, IL-1α, IL-6, TNF-α, E-selectin, tissue factor and endothelin-1 (38). The AGE-RAGE signaling pathway is associated with vascular stiffness and atherosclerosis, as well as adversely affecting endothelial cell responses, vascular smooth muscle cells (VSMCs) function, and platelet activity (39). The interaction of these mechanisms can lead to CMDs and accelerate the development of MVA. Further, STDP could act against MVA by affecting the expression of genes and by attenuating endothelial cell injury, oxidative stress injury, smooth muscle dysfunction, coronary microvascular embolism, and inflammatory injury, as shown by the GO and KEGG enrichment analyses.
We performed PPI analysis of the 138 targets intersected by STDP against MVA, and the core targets were further obtained using the plug-in cytoHubba. The results indicated that IL6, AKT1, STAT3, JUN, IL-1β, PTGS2, CXCL8, CASP3, MMP9, MAPK3, TLR4, CCL2, and VEGFA play critical roles in the PPI network. The IL-6/JAK/STAT3 signaling pathway is closely linked to the inflammatory response (40). STAT3 acts broadly and regulates the expression of over 1,000 gene products. In general, regardless of its subcellular localization, the regulation of autophagy is mainly characterized by the promotion of survival (41). TLR4 is recognized by ligands, such as lipopolysaccharides, to produce inflammatory chemokines and proinflammatory factors that induce inflammatory responses and modulate natural immunity (42). A study by Su et al. (43) showed that the inhibition of the TLR4/MyD88/NF-κB signaling pathway was effective in ameliorating CME-induced myocardial injury chiefly by reducing myocardial inflammatory injury. In addition, total tanshinone, the main active component of STDP, inhibited the expression of lipopolysaccharide-induced inflammatory factors, such as iNOS, COX-2, TNF-α, IL-6, and IL-1β (14). Furthermore, the expression of human coronary artery smooth muscle cell-derived inflammatory mediators, such as CCL2, CXCL8, IL6, and IL-1β, can be suppressed by cardiac protein-associated transcription factors (44). The c-Jun protein is encoded by the gene JUN. The c-Jun constitutes the activator protein 1 (AP-1) early response transcription factor in the form of a dimeric complex and regulates gene expression and cellular function in response to various extracellular and intracellular stimuli (45). The JUN kinase delays Caspase-9 activation via interaction with apoptotic vesicles, thereby promoting cell survival during oxidative stress bursts (46). Vascular endothelial growth factor (VEGF) is a homodimeric vasoactive glycoprotein that plays a crucial role in angiogenesis. On the one hand, VEGFA promotes angiogenesis and increases the density of the microvasculature in the infarct area. On the other hand, it promotes angiogenesis while accelerating plaque growth and increasing plaque instability, thus leading to intraplaque hemorrhage and aggravating the local inflammatory response. Moreover, VEGFA plays a key role in wound healing, mechanical stress, and cytokine stimulation (47). According to a recent study, MMP9 might promote angiogenesis by inhibiting the secretion of vascular endothelial inhibitors via unknown mechanisms (48). Also named COX, PTGS is an enzyme that mediates the conversion of arachidonic acid to PGH2 (49). It has been shown that the expression of PTGS2 is positively correlated with the severity of atherosclerotic lesions (50).
The results of molecular docking showed that some key compounds of STDP are capable of binding well with the corresponding target proteins. The findings revealed that the core target IL-6 receptor demonstrated the strongest binding affinity to tanshinone IIA and that the AKT1 receptor had the strongest affinity to luteolin. STAT3 receptor was close to the binding energy of cryptotanshinone and ginsenoside Rg7, and JUN showed the strongest binding energy to dihydrotanshinone I. The binding energies of IL-1β to cryptotanshinone and tanshinone IIA were close, and TLR4 presented the strongest binding affinity to cryptotanshinone. All these compounds contained several hydrogen bond acceptor donors, which aided them in forming more stable hydrogen bonds with the target protein receptors. This, in turn, enabled the small molecule compound ligands to bind stably to the active site of the corresponding protein. In addition, hydrophobic interactions and van der Waals forces were present between the compound ligands and the protein active residues, and conjugated structures were formed. These forces can effectively immobilize the small molecules in the active center, thus enhancing the stability of the compound in the active pocket of the protein.
The results of our research imply that STDP exhibits multicomponent, multitargeted, multi-pathway, and multilinked regulatory characteristics against MVA. The therapeutic effects of STDP may be exerted via processes such as anti-inflammation, promotion of smooth muscle cell proliferation and differentiation, lipid metabolism, immunomodulation, and regulation of cellular autophagy.
These potential targets and pathways predicted by network pharmacology tools and bioinformatic techniques need to be confirmed by further experimental evidence due to the limitations of network pharmacology. Although STDP has been widely used in the cardiovascular system, its underlying mechanisms need to be further validated experimentally. Furthermore, based on the advantages of STDP as a TCM, research should not be limited to single system related diseases. In the future, studies on STDP should be used for the treatment of multiple systemic diseases, as well as to verify its potential mechanisms through experiments.
Acknowledgments
The authors would like to thank all the reviewers for their valuable comments.
Funding: The research was supported by the National Natural Science Foundation of China (No. 81960079) and the Guangxi Natural Science Foundation (No. 2020GXNSFFA297002).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3976/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3976/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Aldiwani H, Mahdai S, Alhatemi G, et al. Microvascular Angina: Diagnosis and Management. Eur Cardiol 2021;16:e46. [Crossref] [PubMed]
- Jarczewski J, Jarczewska A, Boryczko A, et al. Microvascular angina (Cardiac Syndrome X) from a historical overview, epidemiology, pathophysiology to treatment recommendations - a minireview. Folia Med Cracov 2021;61:95-114. [PubMed]
- Sedlak T, Izadnegahdar M, Humphries KH, et al. Sex-specific factors in microvascular angina. Can J Cardiol 2014;30:747-55. [Crossref] [PubMed]
- Saito Y, Nishi T, Kato K, et al. Resistive reserve ratio and microvascular resistance reserve in patients with coronary vasospastic angina. Heart Vessels 2022;37:1489-95. [Crossref] [PubMed]
- Mahtani AU, Padda IS, Bhatt R. Cardiac Syndrome X. StatPearls; 2022.
- Villano A, Lanza GA, Crea F. Microvascular angina: prevalence, pathophysiology and therapy. J Cardiovasc Med (Hagerstown) 2018;19:e36-9. [Crossref] [PubMed]
- Rakhimov K, Gori T. Non-pharmacological Treatment of Refractory Angina and Microvascular Angina. Biomedicines 2020;8:285. [Crossref] [PubMed]
- Padro T, Manfrini O, Bugiardini R, et al. ESC Working Group on Coronary Pathophysiology and Microcirculation position paper on 'coronary microvascular dysfunction in cardiovascular disease'. Cardiovasc Res 2020;116:741-55. [Crossref] [PubMed]
- Vancheri F, Longo G, Vancheri S, et al. Coronary Microvascular Dysfunction. J Clin Med 2020;9:2880. [Crossref] [PubMed]
- Lin S, Chu J, Zhang L, et al. Protective effects of Shexiang Tongxin Dropping Pill on pituitrin induced acute myocardial ischemia in rats. Mol Med Rep 2017;16:3125-32. [Crossref] [PubMed]
- Zhang Y, Zhao J, He Z, et al. Shexiang Tongxin Dropping Pill Improves Peripheral Microvascular Blood Flow via Cystathionine-γ-Lyase. Med Sci Monit 2019;25:6313-21. [Crossref] [PubMed]
- Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:13. [Crossref] [PubMed]
- Fang S, Dong L, Liu L, et al. HERB: a high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Res 2021;49:D1197-206. [Crossref] [PubMed]
- Gao H, Liu X, Sun W, et al. Total tanshinones exhibits anti-inflammatory effects through blocking TLR4 dimerization via the MyD88 pathway. Cell Death Dis 2017;8:e3004. [Crossref] [PubMed]
- Zhang T, Jin W, Yang S, et al. Study of compositions of musks in different types secreted by forest musk deer (Moschus berezovskii). PLoS One 2021;16:e0245677. [Crossref] [PubMed]
- Yuan R, Huang L, Du LJ, et al. Dihydrotanshinone exhibits an anti-inflammatory effect in vitro and in vivo through blocking TLR4 dimerization. Pharmacol Res 2019;142:102-14. [Crossref] [PubMed]
- Xu X, Zhang W, Huang C, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci 2012;13:6964-82. [Crossref] [PubMed]
- Gfeller D, Grosdidier A, Wirth M, et al. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res 2014;42:W32-8. [Crossref] [PubMed]
- Safran M, Chalifa-Caspi V, Shmueli O, et al. Human Gene-Centric Databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res 2003;31:142-6. [Crossref] [PubMed]
- Amberger JS, Bocchini CA, Scott AF, et al. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res 2019;47:D1038-43. [Crossref] [PubMed]
- Thorn CF, Klein TE, Altman RB. PharmGKB: the pharmacogenetics and pharmacogenomics knowledge base. Methods Mol Biol 2005;311:179-91. [Crossref] [PubMed]
- Piñero J, Queralt-Rosinach N, Bravo À, et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford) 2015;2015:bav028. [Crossref] [PubMed]
- Chen X, Ji ZL, Chen YZ. TTD: Therapeutic Target Database. Nucleic Acids Res 2002;30:412-5. [Crossref] [PubMed]
- Demchak B, Otasek D, Pico AR, et al. The Cytoscape Automation app article collection. F1000Res 2018;7:800. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- von Mering C, Huynen M, Jaeggi D, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 2003;31:258-61. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. [Crossref] [PubMed]
- Burley SK, Berman HM, Kleywegt GJ, et al. Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. Methods Mol Biol 2017;1607:627-41. [Crossref] [PubMed]
- Kim S, Chen J, Cheng T, et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res 2021;49:D1388-95. [Crossref] [PubMed]
- O'Boyle NM, Banck M, James CA, et al. Open Babel: An open chemical toolbox. J Cheminform 2011;3:33. [Crossref] [PubMed]
- Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des 2010;24:417-22. [Crossref] [PubMed]
- Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 2009;30:2785-91. [Crossref] [PubMed]
- Su Q, Li L, Sun Y, et al. Effects of the TLR4/Myd88/NF-κB Signaling Pathway on NLRP3 Inflammasome in Coronary Microembolization-Induced Myocardial Injury. Cell Physiol Biochem 2018;47:1497-508. [Crossref] [PubMed]
- Zhao R, Liu P, Song A, et al. Network pharmacology study on the mechanism of Qiangzhifang in the treatment of panic disorder. Ann Transl Med 2021;9:1350. [Crossref] [PubMed]
- Shimokawa H, Suda A, Takahashi J, et al. Clinical characteristics and prognosis of patients with microvascular angina: an international and prospective cohort study by the Coronary Vasomotor Disorders International Study (COVADIS) Group. Eur Heart J 2021;42:4592-600. [Crossref] [PubMed]
- Tunc E, Eve AA, Madak-Erdogan Z. Coronary Microvascular Dysfunction and Estrogen Receptor Signaling. Trends Endocrinol Metab 2020;31:228-38. [Crossref] [PubMed]
- Rong LL, Yan SF, Wendt T, et al. RAGE modulates peripheral nerve regeneration via recruitment of both inflammatory and axonal outgrowth pathways. FASEB J 2004;18:1818-25. [Crossref] [PubMed]
- Barlovic DP, Thomas MC, Jandeleit-Dahm K. Cardiovascular disease: what's all the AGE/RAGE about? Cardiovasc Hematol Disord Drug Targets 2010;10:7-15. [Crossref] [PubMed]
- Kosmopoulos M, Drekolias D, Zavras PD, et al. Impact of advanced glycation end products (AGEs) signaling in coronary artery disease. Biochim Biophys Acta Mol Basis Dis 2019;1865:611-9. [Crossref] [PubMed]
- Yeung YT, Aziz F, Guerrero-Castilla A, et al. Signaling Pathways in Inflammation and Anti-inflammatory Therapies. Curr Pharm Des 2018;24:1449-84. [Crossref] [PubMed]
- You L, Wang Z, Li H, et al. The role of STAT3 in autophagy. Autophagy 2015;11:729-39. [Crossref] [PubMed]
- Jiao L, Wang CC, Wu H, et al. Copper/zinc-loaded montmorillonite influences intestinal integrity, the expression of genes associated with inflammation, TLR4-MyD88 and TGF-β1 signaling pathways in weaned pigs after LPS challenge. Innate Immun 2017;23:648-55. [Crossref] [PubMed]
- Su Q, Lv X, Sun Y, et al. Role of TLR4/MyD88/NF-κB signaling pathway in coronary microembolization-induced myocardial injury prevented and treated with nicorandil. Biomed Pharmacother 2018;106:776-84. [Crossref] [PubMed]
- Liu L, Bankell E, Rippe C, et al. Cell Type Dependent Suppression of Inflammatory Mediators by Myocardin Related Transcription Factors. Front Physiol 2021;12:732564. [Crossref] [PubMed]
- Yu B, Miao ZH, Jiang Y, et al. c-Jun protects hypoxia-inducible factor-1alpha from degradation via its oxygen-dependent degradation domain in a nontranscriptional manner. Cancer Res 2009;69:7704-12. [Crossref] [PubMed]
- Tran TH, Andreka P, Rodrigues CO, et al. Jun kinase delays caspase-9 activation by interaction with the apoptosome. J Biol Chem 2007;282:20340-50. [Crossref] [PubMed]
- Braile M, Marcella S, Cristinziano L, et al. VEGF-A in Cardiomyocytes and Heart Diseases. Int J Mol Sci 2020;21:5294. [Crossref] [PubMed]
- Gupta A, Zhou CQ, Chellaiah MA. Osteopontin and MMP9: Associations with VEGF Expression/Secretion and Angiogenesis in PC3 Prostate Cancer Cells. Cancers (Basel) 2013;5:617-38. [Crossref] [PubMed]
- Lee HJ, Kim SR, Jung YJ, et al. Cyclooxygenase-2 induces neoplastic transformation by inhibiting p53-dependent oncogene-induced senescence. Sci Rep 2021;11:9853. [Crossref] [PubMed]
- Zhou Y, Zhou H, Hua L, et al. Verification of ferroptosis and pyroptosis and identification of PTGS2 as the hub gene in human coronary artery atherosclerosis. Free Radic Biol Med 2021;171:55-68. [Crossref] [PubMed]
(English Language Editor: J. Jones)


