Microbiome and metabolome analysis to clarify the interaction between the urine microbiota and serum metabolites in Chinese patients with immunoglobulin A nephropathy
Highlight box
Key findings
• We verified the connection between the disruption of the microbiota and serum metabolites, along with the clinical parameters, in patients with IgAN.
What is known and what is new?
• A relational network between gut microbiota and serum metabolites in IgAN was found. Altered fecal microbiota has also been identified as a possible tool to distinguish between patients with IgAN and HCs.
• While previously considered sterile, the UT now hosts a range of bacteria in healthy individuals. We observed a shift in the abundance of bacteria and metabolites in patients with IgAN. Our study clarified the relationship between serum metabolites, urine microbiota, and disease in those with IgAN.
What is the implication, and what should change now?
• This will help to provide further research and develop new tools for preventing and delaying the occurrence of the disease.
Introduction
Immunoglobin A nephropathy (IgAN) is recognized as the most common form of primary chronic glomerulonephritis worldwide. IgAN is responsible for approximately 45.26% of total primary glomerular disease cases in China. About 10–20 years after diagnosis, one-third of patients with IgAN progress to end-stage renal disease (ESRD). These patients require renal replacement therapies (including hemodialysis or peritoneal dialysis or transplantation) for survival (1-3). Currently, an accurate diagnosis of IgAN depends almost completely on percutaneous renal biopsies (4). Meanwhile, IgAN’s etiology and pathogenesis remain unknown but it is generally accepted that the deposition of galactose-deficient IgA1 (Gd-IgA1) (5). A previous study has found that genes, environment, living habits and patterns play an important role in the pathogenesis of IgAN (6) and that systematic changes in cellular metabolism are caused by these intrinsic and extrinsic factors. In recent studies, some metabolites, such as urinary glycine, have been shown to be protective biomarkers for IgAN (7,8). Moreover, the alteration of fatty acids and amino acids has been shown to activate the unique metabolic pathway of IgAN (9). These metabolites were found to be changed in connection with changes in the microbiome and discovered because of the recent rise of gut/kidney axis research. Altered fecal microbiota has also been identified as a possible tool to distinguish between patients with IgAN and healthy controls (HCs) (10). An increasing amount of research points to the close relationship between microbiota and metabolites as being critical in the occurrence and development of diseases. IgAN therapy currently depends on treatments supported by only low-level evidence, mainly relating to immunosuppressants and the renin-angiotensin system (RAS). Emerging research, however, has reported that probiotic supplements such as Lactobacillus (11) and polyunsaturated fatty acids are integral to both the prevention and treatment of IgAN.
While previously considered sterile, the urinary tract (UT) now hosts a range of bacteria in healthy individuals (12). At present, there are many studies related to gut microbiota, but we currently know little about the role of UT microbiota and its mechanism. Several studies have found specific bacterial communities in the healthy UT based on advances in molecular biology techniques (13,14), and we can observe changes in the UT microbiome in some urinary system diseases, such as chronic pelvic pain syndrome, urologic cancers, neurogenic bladder dysfunction and urinary incontinence (12,13). The network of the urinary microbiome and the serum metabolome has not thus far been analyzed, and yet the close relationship between microbiota and metabolites may likely have considerable relevance for therapy. This study thus aimed to clarify the network of different serum metabolites and urinary bacteria and to provide some means for clinical diagnosis and treatment. We present the following article in accordance with the MDAR checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5334/rc).
Methods
Patient information and sample collection
A total of 14 patients diagnosed with IgAN according to renal biopsies and 15 healthy people were recruited at the First Affiliated Hospital of Jinan University. These participants were informed of the study and signed an informed consent form. The basic clinical information of the participants, including gender, age, body mass index (BMI), fasting blood glucose, hypertension, liver and renal functions, and pathological data of IgAN, were recorded using the Oxford classification system. Patients were excluded if they had malignant tumors, infectious diseases, diabetes mellitus, severe liver dysfunction, cardiac disease, other immune diseases, a history of excessive alcohol intake, or received any anti-inflammatory or probiotics treatment within the past 3 months (15). Participants in the HCs group were also excluded if they had a history of autoimmune disease.
Serum and urine samples from all volunteers were collected on the morning before a recent renal biopsy. Before collection, the volunteers had to thoroughly wash their hands and clean their external genital area with water and soap. Women washed the area around the external urethral orifice and vaginal introitus, then dipped 4 cotton swabs in iodophor, and moved them back and forth to wipe the genitals. The men were asked to disinfect the area around the urethral orifice after the foreskin had been withdrawn and the glans of the penis exposed. Without interrupting the micturition, the volunteers urinated into the toilet and then into a sterile container. The self-collected midcourse clean urine specimens were quickly transferred to the biological sample laboratory. These samples were stored in sterile Eppendorf (EP) tubes at –80 ℃. Blood samples were taken in the morning after volunteers had fasted all night. Volunteers with hemolysis present in their serum samples were excluded from the study. The collected supernatants, which were centrifuged at 3,000 rpm at 4 ℃ for 10 minutes, were then transferred to a –80 ℃ refrigerated cabinet for long-term storage. This study was authorized and reviewed by the institutional review board of the First affiliated Hospital of Jinan University (No. KY-2020-034) and conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Metabolite extraction and liquid chromatography-tandem mass spectrometry analysis
A total of 400 µL of extract solution (acetonitrile: methanol =1:1; containing isotopic labeled internal standard mixture) was added to the 100 µL sample in an EP tube. After the samples were rotated for 30 s, we sonicated them in an ice-water bath for 10 min and cultured them at –40 ℃ for 1 h to precipitate. The samples were then centrifuged at 12,000 rpm at 4 ℃ for 15 min. We collected the resulting supernatant in a clean bottle for analysis. We then mixed the aliquots from all samples and analyzed the samples with liquid chromatography-tandem mass spectrometry (LC-MS/MS; UHPLC system with UPLC BEH amide column and Q Exactive HFX mass spectrometer, Thermo Fisher Scientia) and subjected them to multivariate analysis. The detailed experimental methods we used are outlined in previous papers published by our research group (9).
Metabolomics data preprocessing and annotation
The original data was converted to the mzXML format using ProteoWizardsoftware. The peak extraction, calibration, and integration based on XCMS were performed using an internal program written in R. A further processing of the data matrix was carried out by removing more than 50% of the peaks of the missing values in the samples using half of the minimum of the simulation method as a simulation and filling in some gaps in the data.The new data matrices were standardized by internal standards, and 3 databases were applied to metabolite annotation: an in-house MS2 database (BiotreeDB), the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (www.genome.jp/kegg), and the Human Metabolome Database (HMBD) online database (www.hmdb.ca). The cutoff value for comments was set as 0.3. We used a multivariate statistical analysis that included a principal component analysis and an orthogonal projection to latent structures discriminant analysis (OPLS-DA) to compare HCs and IgAN metabolomic characteristics.
Urine DNA extraction and 16S sequencing
The Mobio Powersoil DNA Isolation Kit (Qiagen) was used to extract genomic DNA from urine samples according to the manufacturer's instructions. The V3 and V4 regions of the 16S ribosomal RNA (rRNA) genes were amplified with the following primers: F-primer: 5'-ACTCCTACGGGAGGCAGCA-3'; and R-primer: 5'-GGACTACHVGGGTWTCTAAT-3'. With the Illumina HiSeq, we analyzed the samples using a paired-end sequencing strategy after purifying the polymerase chain reaction products using ampoule XP magnetic beads (Beckman Coulter, UK).
Sequencing data analysis
Filtered paired-end reads were modified using Trimmomatic v. 0.33 software (Illumina, USA) (16). Using Cutadapt (version 1.9.1), the merging primer sequences were identified and removed. The reads were next combined using FLASH 1.2.11 software (17), and the chimeric sequences were removed using UCHIME 8.1 (18). The high-quality sequences that were obtained were then used for subsequent analysis. USEARCH software version 10.0 classified sequences with a similarity level of 97% as operational taxonomic units (OTUs) (19). We then annotated the taxonomic information based on the Silva database (20), and different phylogenetic levels (phylum, class, order, family, genus, and species) were assigned to the OTUs. We used QIME2 (https://qiime2.org/) to analyze alpha diversity and the Wilcoxon rank sum test to compare the amount of data and statistical difference diversity. In order to identify the bacterial taxa whose sequences were differentially abundant between the IgAN and control groups, we used linear discriminant analysis (LDA) and effect size (LEfSe). All sequencing data can be found in the National Center for Biotechnology Information (NCBI) database (accession No. SUB11247081).
Statistical analysis
We used the Student’s t-test and the Wilcoxon rank sum test to conduct the differential analysis of the measured data. We assessed the correlations between UT bacteria and serum metabolites using Spearman correlation analysis and found a correlation between the microbiome and metabolome and clinical indicators. Statistical calculations were performed with the SPSSAU project, an online software application (https://www.spssau.com). Metabolites were identified that had a variable importance in the project (VIP) >1 and a P value <0.05 that indicated a significant difference. The plot was optimized using GraphPad Prism 9 (GraphPad Software) and the OmicStudio tools (https://www.omicstudio.cn/tool). We calculated the specificity and sensitivity of the core metabolites, microbiome, and clinical symptoms using the receiver operating characteristic (ROC) curve.
Results
Summary of clinical characteristics
A total of 29 participants, including 14 patients with IgAN (mean age 36.50±13.83 years) and 15 HCs (mean age 31.00±12.14 years), were enrolled in the study at the First Affiliated Hospital of Jinan University. We used independent sample t-tests to compare the two groups. There was a statistical difference in the serum albumin, 24-h proteinuria, full blood count, urea nitrogen, creatine, and uric acid (P<0.05) between the two groups. However, no significant differences were found in age, BMI, alanine aminotransferase (ALT), or aspartate transaminase (AST). Although the systolic blood pressure of those with IgAN was in the normal range (90–130 mmHg) according to the guidelines of the American College of Cardiology (ACC) and the American Heart Association (AHA), it was significantly higher than that of the HCs group. Detailed descriptions of the IgAN patients and HCs can be found in Tables S1,S2.
OTUs and diversity analyses in bacteria
The urinary bacteria of 14 patients with IgAN and 15 HCs were analyzed using 16S rRNA gene sequencing. In this study, we obtained an average of 74,399 valid tags (average length: 418 bp) and 2,157,579 total reads. We then identified 1,464 OTUs in the HC group and 1,466 OTUs in the IgAN group, including 1,463 shared OTUs, indicating a 97% similarity between the two groups (Figure 1A). The sequencing samples obtained were sufficient for further taxon identification on the basis of the rarefaction curve (Figure 1B) and species accumulation curve (Figure 1C). We found no significant difference in species diversity (Shannon and Simpson indices) or species richness [Chao1 and abundance-based coverage estimator (ACE)] in the Student’s t tests.

We also identified 10 main bacterial phyla (Figure 1D), among which Firmicutes, Bacteroidetes, Proteobacteria, Acidobacteria, Actinobacteria, and Verrucomicrobia constituted over 93% of the bacteria in all 16S rRNA sequence groups. Our data also showed that the relative abundance of Acidobacteria was higher in patients with IgAN (control: 9%; IgAN: 11%; P=0.04; Figure 1E), and that Lactobacillus (control: 16%; IgAN: 10%) levels decreased in the IgAN group at the genus level according to the Wilcoxon rank sum test (P=0.00776183; Figure 1F).
We used a Metastats analysis to predict the core microbiota at different levels of bacterial classification between the IgAN and control groups (P<0.05; LDA >4; Figure 2A; Table 1) and cladograms to describe the evolutionary relationship between microorganisms based on LDA >4 (Figure 2B). The results showed that the abundance of the phylum Acidobacteria increased in patients with IgAN. By contrast, patients with IgAN exhibited a loss of Lactobacilli (order: Lactobacillales; family: Lactobacillaceae; genus: Lactobacillus) and Bacilli at the class level. In summary, the microbiome of patients with IgAN compared with HCs expressed a disorder of the microbial community which may lead to the disorder of immune mechanisms (21).

Table 1
| Bacteria | Metastats analysis | Wilcoxon test | LEfSe analysis | ||
|---|---|---|---|---|---|
| HC (mean) | IgAN (mean) | HC (LDA value) | IgAN (LDA value) | ||
| Phylum | |||||
| Acidobacteria | 8.64% | 10.70% | 0.04024279 | 0.036180617 | 4.00270126 |
| Class | |||||
| Bacilli | 17.50% | 12% | 0.04468533 | 4.547905826 | 0.04468533 |
| Order | |||||
| Lactobacillales | 17.20% | 11.60% | 0.04024279 | 4.551476082 | 0.04024279 |
| Family | |||||
| Lactobacillaceae | 15.70% | 10.10% | 0.00776183 | 4.544844194 | 0.00776183 |
| Genus | |||||
| Lactobacillus | 15.70% | 10.10% | 0.00776183 | 4.543791671 | 0.00776183 |
LEfSe, linear discriminant analysis effect size; HC, healthy control; IgAN, immunoglobin A nephropathy; LDA, linear discriminant analysis.
Changes of serum metabolites in patients with IgAN
We performed an LC-MS/MS analysis of the serum metabolite alterations in patients with IgAN (VIP >1; P<0.05), and found 168 differential metabolites (Table S3), divided into several classes, such as peptides, lipids, amino acids, and carbohydrates. Amino acids, peptides, organic heterocyclic compounds, and lipids accounted for the majority of major alterations in the differential metabolites of IgAN. We also found that amino acids and peptides showed an upward trend (Figure 3A), while lipids and lipid-like molecules in the serum metabolites of patients with IgAN showed a descending trend (Figure 3B). Cellobiose, homocitrulline, apo-[3-methylcrotonoyl-CoA:carbon-dioxide ligase (ADP-forming)], prolylhydroxyproline, 24-epibrassinolide, butyrylcarnitine, guanidinosuccinic acid, formiminoglutamic acid, and diethanolamine were the metabolites with a VIP >2 and log fold change >2, and thus were suspected to be the core metabolites in patients with IgAN (Figure 3C). In addition, we found that the histidine metabolism pathway was statistically different (P<0.05; impact value >0.1) through an enrichment analysis in Metaboanalyst v. 5.0 (Figure 3D).

Correlation between altered microbiome and metabolism
We clarified the relationship between the microbiome and differential metabolites through a Spearman correlation analysis (Figure 4A) and found that 12 different metabolites were negatively correlated with Lactobacillus and 19 different metabolites were positively correlated with Lactobacillus (Table S4). Interestingly, Lactobacillus, which was positively correlated with guanidinoacetic acid, has a negative correlation with apo-[3-methylcrotonoyl-CoA:carbon-dioxide ligase (ADP-forming)] and diethanolamine in core metabolites (r>0.5; P<0.05). The disturbance of Lactobacillus number may affect the changes of these metabolites.

Correlation analysis between metabolites, microbiome, and clinical symptoms
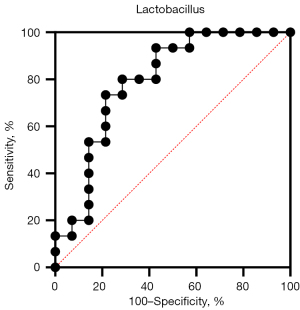
Using Spearman correlation analysis, we assessed the relationship between metabolites, microbiome, and clinical indicators (Figure 4B). We found that some core metabolites including homocitrulline, apo-[3-methylcrotonoyl-CoA:carbon-dioxide ligase (ADP-forming)], butyrylcarnitine, formiminoglutamic acid (FIGLU), diethanolamine, and prolylhydroxyproline had a positive correlation with mili-total protein (MTP), whereas cellobiose, butyrylcarnitine, and prolylhydroxyproline had a negative correlation with serum albumin, with 24-epibrassinolide being negatively correlated with MTP (P<0.05; |rho| >0.5). Lactobacillus was also negatively correlated with MTP and urea nitrogen but positively correlated with serum albumin (Table S5). There was also no significant difference in core metabolites when the Oxford classification of IgAN was used. Lactobacillus and core metabolites were determined to be predictors of IgAN by an area under the curve (AUC) analysis. Lactobacillus showed an excellent prediction ability for the diagnosis of IgAN in the ROC curve (AUC >0.79; 95% CI: 0.6176–0.9633; Figure 5).
Discussion
Interactions between genetics and the environment are thought to determine IgAN’s development (22,23). The microbiome is currently recognized as a key environmental factor that contributes to the development and progression of the disease. While sequencing adds the comprehensive understanding of microorganisms, the metabolomics based on mass spectrometry is a pivotal technology of detect small molecules produced by microbiome (24). Through the correlation analysis of microbiome and metabolomics, we can provide broader ideas and information for the screening of disease markers. Most existing multiomics studies elaborate on changes in the serum metabolome and gut microbiome, and a few studies have tried to examine the alterations in the serum metabolome and urinary microbiome. It has long been thought that the UT was a sterile environment, but the introduction of new high-throughput sequencing technologies and improved culture schemes for microbiome research has proven that the UT is not sterile (12,25,26). It has also been reported that Limosilactobacillus urinaemulieris sp. nov. and Limosilactobacillus portuensis sp. nov. are present in the urine of healthy women (27). The complicated microbial communities of the human UT can now be analyzed using high-throughput molecular sequencing of bacterial 16S rRNA genes (28). Recent studies have reported that the bacterial taxa of the urinary microbiome have an important influence on diseases and homeostasis in the UT (29-31). For example, some studies indicated an association between lower urinary tract symptoms (LUTS), or incontinence, and the human urinary microbiome (32,33). The dysbiosis of several key urinary bacteria has also been found to be related to interleukin-8 (IL-8) in type 2 diabetes mellitus (T2DM) (34). We hypothesized that urinary microbiota and metabolites might also be a feasible detection tool for IgAN.
Based on alpha and beta diversity analyses, we found that bacterial species did not differ significantly between the IgAN group and the HC group, which is similar to findings in previous research (9). We also found that the amount and diversity of bacteria was different in patients with IgAN. It has been reported that the main bacterial taxa of healthy people (aged 22–51 years) are Lactobacillus, Aerococcus, Klebsiella, Staphylococcus, Corynebacterium, Gardnerella, Streptococcus, Escherichia, Prevotella, and Enterococcus (35). We found that the distributions of the major phyla in the UT (Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria) were roughly consistent with the results of previous human adult gut studies (36-38). There was a 1.24-fold increase in Acidobacteria at the phylum level (P<0.05), a 1.9-fold increase in Actinomyces in the genus level (P<0.05), and a 2.6-fold decrease in Lactobacillaceae (P<0.05), but a 0.34-fold decrease in Lactobacillus (P<0.05) in our Wilcoxon rank sum test, which indicated that the specific species of urinary microbiota in patients with IgAN was altered. Our Metastats analysis found that Actinobacteria was more abundant at the phylum level in patients with IgAN and that Actinotignum was a notable differential microorganism at the genus level (P<0.01). A previous study using both 16S DNA and rRNA found that Actinobacteria of the main bacterial phyla significantly differed in the fecal microbiota of patients with IgAN (10). A study revealed that Actinotignum, known for its proinflammatory features, could be an early polybacterial biofilm colonizer. Actinotignum usually presents in the UT as an opportunistic pathogen and coagent of various polymicrobial infections, but it has not been found in feces (39). Studies have also shown that Actinotignum could be associated with noninfectious conditions and diseases, such as chronic inflammation, prostatism, and bladder cancer (31,40-43). Therefore, we speculate that the increase of Actinomyces in urinary flora may be related to IgAN.
We found that Lactobacillus at the genus level was lower in patients with IgAN compared to healthy controls, which is in agreement with a previous study that found that Lactobacillus genera to be higher in controls and Lactobacillaceae to be lower in patients with IgAN (P<0.05) (10). It has also been reported that Lactobacillus can reduce inflammation of the kidney and also reduce injury in renal tubular epithelial cells. Lactobacillus may also protect the kidney through an independent mechanism against the interference of the original bacterial flora. Lactobacillus can decrease oxidative stress, reduce the proinflammatory response, and increase kidney function in immune responses. We speculate that Lactobacillus supplementation may work as a preventive approach against chronic kidney disease (CKD) (44).
Metabolites play an important role in the development and progression of renal diseases as mediators of bacterial functional activities (9). Previous studies have reported the alteration of amino acids, such as Met, Glu, and Pro, in the serum samples of patients with IgAN (45,46). This finding is in line with our research, which showed both increased differential metabolites (amino acids, oligopeptides, and amines) and decreased differential metabolites (medium-/long-chain fatty acids; Figure 3A). We hypothesize that the level of amino acids in serum was changed through the alteration of the tricarboxylic acid cycle and the increased hydrolysis of proteins in cell necrosis (10). We found that an increase of oligopeptides, rather than polypeptides, demonstrated bacterial proteolytic fermentation. Several studies have reported that protein assimilation, such as digestion and absorption, are impaired in patients with ESRD, suggesting that oligopeptides are more easily absorbed due to noncompetitive transport and low energy consumption (47-49). In addition, we found a decrease in lipids, especially in unsaturated fatty acids (e.g., arachidonic acid). Studies have shown that long-chain polyunsaturated fatty acids (LCPUFA) play a preventive and deferring role in renal patients with cardiovascular disease (CVD), and that LCPUFA supplementation can extend the life of patients on long-term dialysis (50,51). A low dose of LCPUFA can also help lower estimated glomerular filtration rate (eGFR) (52). In our study, we found changes in differential metabolites in the serum samples of patients with IgAN as compared to healthy controls.
A previous study reported that the dysbiosis of urinary microbiota is related to proinflammatory chemokine IL-8 (34). Here, we studied the relationship between serum metabolites and urinary microbiota (Figure 4A). We found that some serum metabolites, such as trimethylamine-N-oxide (TMAO) and D-mannose, were related to Lactobacillus (P<0.05; |rho| >0.5). Several researchers reported that atherosclerosis, mediated by TMAO, could be an important cause of kidney and heart disease from dietary phosphatidylcholine (53). TMAO concentration is also associated with eGFR, prognosis, and even long-term survival in CKD (54,55). Some probiotics, such as Lactobacillus, have been shown to inhibit the synthesis of TMAO, reducing inflammatory signaling and improving renal function in CKD (56,57). Some studies suggest that D-mannose can inhibit bacterial adherence to uroepithelial cells (58,59). We speculate that Lactobacillus and D-mannose may work together to aid urethral immunity. KEGG analysis showed that histidine pathway was enriched in the serum of patients with IgAN. The microbiota can produce L-histidine through the histidine metabolic pathway, which is a potential biomarker of septic acute kidney injury (AKI) (60).
In this study, Spearman correlation analysis indicated a correlation between core metabolites, the microbiome, and clinical indicators in patients with IgAN. Similarly, homocitrulline, involved in CKD progression, has been found to be related to MTP (61); Butyrylcarnitine, which is positively correlated with MTP, was also shown to be a predictive biomarker in the diagnosis of renal cell carcinoma (RCC) (62); meanwhile, prolylhydroxyproline, which has also been positively correlated with MTP, may be a marker for cognitive impairment in patients receiving long-term maintenance dialysis (63). We also found that Lactobacillus was negatively correlated with urea nitrogen and MTP but positively correlated with serum albumin. This is consistent with previous studies which found that some Lactobacilli can reduce urea nitrogen and upregulate albumin levels (64,65). Our research still needs to be verified with larger samples, but using an ROC model, we found that Lactobacillus is a good predictor of IgAN (AUC >0.79; Figure 5).
Urine, a waste product, is created through the various metabolic endpoints, which eventually reach the bladder and kidneys. Urethral microbiota are affected by external factors, such as lifestyle, diet, and the environmental, and has ample time to interact with and alter these metabolisms filtered by blood through kidneys (7,31). Urethral microbiota affects the kidney through a host of mechanisms in this process, such as evading the protective factors of the host or inhibiting host IgA transport (66). Moreover, a study on sterile mice have shown that microbial deficiency is associated with an impaired immune system and behavioral or neurological diseases (67). Therefore, the changes in the urethral microbiota of patients with IgAN as compared with healthy controls may increase the risk of alterations in renal pathologies. We found a close relationship between urethral microbiota and serum metabolites. There may be a similar mechanism in the human body that causes the deposition of polymeric and hypogalactosylated IgA1 (Gd-IgA1) in the glomerular mesangium or capillary wall, leading to IgAN. Renal insufficiency changed the composition of serum metabolites, so we hypothesize that urethral microbiota interacts with serum metabolites.
Currently, the main treatments for IgAN are immunosuppressive drugs and systemic corticosteroids, but the minimal therapeutic effects and risk of side effects limit their value and application. Our research may provide a therapeutic approach including targeting microbiota modulation or restoration and maintenance of metabolic balance. A recent study of the identification of urinary microbiota using 16S rRNA and culturomics, proposed that the 64% of species in urinary microbiota, which originates from the gut, overlap with gut microbiota (68). The gut microbiota is helpful in maintaining the diversity of prokaryotes in the UT and this study highlighted reduction in recurrence of urinary tract infections after fecal microbiota transplantation (FMT) (69). Perhaps changing the imbalance of metabolites and microbiota could delay the occurrence of IgAN and reduce the impact of the disease. Some researchers have reported that the preventive supplement of D-Mannose and Lactobacillus effectively prevented UT infection (70-72). Furthermore, adjunctive probiotic supplementation may mitigate the increase of TMAO in proteolytic fermentation and decrease the risk of CVD and dementia (73). A reasonable supplement of probiotics and polyunsaturated fatty acids may be a key tool for preventing and treating IgAN.
Our study had some limitations. Our combined microbiome and metabolome analysis results would be more robust if the urinary differential metabolites had been added to the experiment. The results of our study should also be verified on more samples. Now, with more convenient and faster sequencing technologies, we can detect the size and grandeur of microbial communities. Application of the microbiome and metabolome will become a powerful non-invasive target for precision medicine. However, we still lack understanding of the molecular functions encoded by microbial genes. In addition, the joint analysis of multi-omics will be a major trend. We will use the expanded quantitative urine culture (EQUC) to detect the low-abundance uropathogens or bacteria in the future (74,75). EQUC associates with sequencing tools, such as targeted amplicon sequencing, metagenomic sequencing and long-read sequencing, to further describe the urine microbiota and build an expanded genome sequencing isolate banking.
Conclusions
We observed a shift in the abundance of bacteria and metabolites in patients with IgAN. Our study clarified the relationship between serum metabolites, urine microbiota, and disease in those with IgAN, which will help to provide further research and develop new tools for preventing and delaying the occurrence of the disease.
Acknowledgments
Funding: This work was supported by Guangdong Provincial Special Projects in Key Fields of Colleges and Universities (No. 2021ZDZX2042), the Project of Guangdong Bureau of Traditional Chinese Medicine (No. 20202044), the Teaching Reform Project of Jinan University (No. JG2019043), the Guangdong Postgraduate Education Project (No. 2020XSLT10), the Science and Technology Plan of Shenzhen (No. JCYJ20190807153405508), the Guangzhou Science and Technology Plan (No. a01219), and the Special Projects in Key Fields of Ordinary Colleges and Universities in Guangdong Province (No. 2021ZDZX2042).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5334/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5334/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5334/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was authorized and reviewed by the institutional review board of the First Affiliated Hospital of Jinan University (No. KY-2020-034) and conducted in accordance with the Declaration of Helsinki (as revised in 2013). These patients were informed of the study and signed an informed consent form.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wyatt RJ, Julian BA. IgA nephropathy. N Engl J Med 2013;368:2402-14. [Crossref] [PubMed]
- Li LS, Liu ZH. Epidemiologic data of renal diseases from a single unit in China: analysis based on 13,519 renal biopsies. Kidney Int 2004;66:920-3. [Crossref] [PubMed]
- Zürbig P, Jerums G, Hovind P, et al. Urinary proteomics for early diagnosis in diabetic nephropathy. Diabetes 2012;61:3304-13. [Crossref] [PubMed]
- Gao J, Wang Y, Dong Z, et al. A novel differential diagnostic model based on multiple biological parameters for immunoglobulin A nephropathy. BMC Med Inform Decis Mak 2012;12:58. [Crossref] [PubMed]
- Suzuki H, Fan R, Zhang Z, et al. Aberrantly glycosylated IgA1 in IgA nephropathy patients is recognized by IgG antibodies with restricted heterogeneity. J Clin Invest 2009;119:1668-77. [Crossref] [PubMed]
- Fan P, Song J, Chen Q, et al. The influence of environmental factors on clinical pathological changes of patients with immunoglobulin A nephropathy from different areas of China. Ren Fail 2018;40:597-602. [Crossref] [PubMed]
- Josephs-Spaulding J, Krogh TJ, Rettig HC, et al. Recurrent Urinary Tract Infections: Unraveling the Complicated Environment of Uncomplicated rUTIs. Front Cell Infect Microbiol 2021;11:562525. [Crossref] [PubMed]
- Park S, Lee J, Yang SH, et al. Comprehensive metabolomic profiling in early IgA nephropathy patients reveals urine glycine as a prognostic biomarker. J Cell Mol Med 2021;25:5177-90. [Crossref] [PubMed]
- Wu H, Tang D, Zheng F, et al. Identification of a novel interplay between intestinal bacteria and metabolites in Chinese patients with IgA nephropathy via integrated microbiome and metabolome approaches. Ann Transl Med 2021;9:32. [Crossref] [PubMed]
- De Angelis M, Montemurno E, Piccolo M, et al. Microbiota and metabolome associated with immunoglobulin A nephropathy (IgAN). PLoS One 2014;9:e99006. [Crossref] [PubMed]
- Lee TH, Park D, Kim YJ, et al. Lactobacillus salivarius BP121 prevents cisplatin-induced acute kidney injury by inhibition of uremic toxins such as indoxyl sulfate and p-cresol sulfate via alleviating dysbiosis. Int J Mol Med 2020;45:1130-40. [Crossref] [PubMed]
- Aragón IM, Herrera-Imbroda B, Queipo-Ortuño MI, et al. The Urinary Tract Microbiome in Health and Disease. Eur Urol Focus 2018;4:128-38. [Crossref] [PubMed]
- Zerdan MB, Moukarzel R, Naji NS, et al. The Urogenital System's Role in Diseases: A Synopsis. Cancers (Basel) 2022;14:3328. [Crossref] [PubMed]
- Jones-Freeman B, Chonwerawong M, Marcelino VR, et al. The microbiome and host mucosal interactions in urinary tract diseases. Mucosal Immunol 2021;14:779-92. [Crossref] [PubMed]
- Yu Z, Zhu Y, Fu J, et al. Enhanced NADH Metabolism Involves Colistin-Induced Killing of Bacillus subtilis and Paenibacillus polymyxa. Molecules 2019;24:387. [Crossref] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. [Crossref] [PubMed]
- Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011;27:2957-63. [Crossref] [PubMed]
- Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194-200. [Crossref] [PubMed]
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 2013;10:996-8. [Crossref] [PubMed]
- Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590-6. [Crossref] [PubMed]
- Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016;16:341-52. [Crossref] [PubMed]
- Li M, Wang L, Shi DC, et al. Genome-Wide Meta-Analysis Identifies Three Novel Susceptibility Loci and Reveals Ethnic Heterogeneity of Genetic Susceptibility for IgA Nephropathy. J Am Soc Nephrol 2020;31:2949-63. [Crossref] [PubMed]
- Suzuki H, Kiryluk K, Novak J, et al. The pathophysiology of IgA nephropathy. J Am Soc Nephrol 2011;22:1795-803. [Crossref] [PubMed]
- Bauermeister A, Mannochio-Russo H, Costa-Lotufo LV, et al. Mass spectrometry-based metabolomics in microbiome investigations. Nat Rev Microbiol 2022;20:143-60. [Crossref] [PubMed]
- Magistro G, Stief CG. The Urinary Tract Microbiome: The Answer to All Our Open Questions? Eur Urol Focus 2019;5:36-8. [Crossref] [PubMed]
- Hall A, Versalovic J. Microbial Metabolism in the Mammalian Gut: Molecular Mechanisms and Clinical Implications. J Pediatr Gastroenterol Nutr 2018;66:S72-9. [Crossref] [PubMed]
- Ksiezarek M, Ribeiro TG, Rocha J, et al. Limosilactobacillus urinaemulieris sp. nov. and Limosilactobacillus portuensis sp. nov. isolated from urine of healthy women. Int J Syst Evol Microbiol 2019; [Crossref] [PubMed]
- Hiergeist A, Gessner A. Clinical implications of the microbiome in urinary tract diseases. Curr Opin Urol 2017;27:93-8. [Crossref] [PubMed]
- Wolfe AJ, Toh E, Shibata N, et al. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol 2012;50:1376-83. [Crossref] [PubMed]
- Siddiqui H, Nederbragt AJ, Lagesen K, et al. Assessing diversity of the female urine microbiota by high throughput sequencing of 16S rDNA amplicons. BMC Microbiol 2011;11:244. [Crossref] [PubMed]
- Whiteside SA, Razvi H, Dave S, et al. The microbiome of the urinary tract--a role beyond infection. Nat Rev Urol 2015;12:81-90. [Crossref] [PubMed]
- Drake MJ, Morris N, Apostolidis A, et al. The urinary microbiome and its contribution to lower urinary tract symptoms; ICI-RS 2015. Neurourol Urodyn 2017;36:850-3. [Crossref] [PubMed]
- Thomas-White KJ, Kliethermes S, Rickey L, et al. Evaluation of the urinary microbiota of women with uncomplicated stress urinary incontinence. Am J Obstet Gynecol 2017;216:55.e1-55.e16. [Crossref] [PubMed]
- Ling Z, Liu F, Shao L, et al. Dysbiosis of the Urinary Microbiota Associated With Urine Levels of Proinflammatory Chemokine Interleukin-8 in Female Type 2 Diabetic Patients. Front Immunol 2017;8:1032. [Crossref] [PubMed]
- Fouts DE, Pieper R, Szpakowski S, et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med 2012;10:174. [Crossref] [PubMed]
- Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480-4. [Crossref] [PubMed]
- Yildirim S, Yeoman CJ, Sipos M, et al. Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS One 2010;5:e13963. [Crossref] [PubMed]
- Alhagamhmad MH, Day AS, Lemberg DA, et al. An overview of the bacterial contribution to Crohn disease pathogenesis. J Med Microbiol 2016;65:1049-59. [Crossref] [PubMed]
- Kotásková I, Syrovátka V, Obručová H, et al. Actinotignum schaalii: Relation to Concomitants and Connection to Patients' Conditions in Polymicrobial Biofilms of Urinary Tract Catheters and Urines. Microorganisms 2021;9:669. [Crossref] [PubMed]
- Beguelin C, Genne D, Varca A, et al. Actinobaculum schaalii: clinical observation of 20 cases. Clin Microbiol Infect 2011;17:1027-31. [Crossref] [PubMed]
- Tschudin-Sutter S, Frei R, Weisser M, et al. Actinobaculum schaalii - invasive pathogen or innocent bystander? A retrospective observational study. BMC Infect Dis 2011;11:289. [Crossref] [PubMed]
- Reinhard M, Prag J, Kemp M, et al. Ten cases of Actinobaculum schaalii infection: clinical relevance, bacterial identification, and antibiotic susceptibility. J Clin Microbiol 2005;43:5305-8. [Crossref] [PubMed]
- Barberis C, Cittadini R, del Castillo M, et al. Actinobaculum schaalii causing urinary tract infections: report of four cases from Argentina. J Infect Dev Ctries 2014;8:240-4. [Crossref] [PubMed]
- Huang H, Li K, Lee Y, et al. Preventive Effects of Lactobacillus Mixture against Chronic Kidney Disease Progression through Enhancement of Beneficial Bacteria and Downregulation of Gut-Derived Uremic Toxins. J Agric Food Chem 2021;69:7353-66. [Crossref] [PubMed]
- Sui W, Li L, Che W, et al. A proton nuclear magnetic resonance-based metabonomics study of metabolic profiling in immunoglobulin a nephropathy. Clinics (Sao Paulo) 2012;67:363-73. [Crossref] [PubMed]
- Ranganathan N, Friedman EA, Tam P, et al. Probiotic dietary supplementation in patients with stage 3 and 4 chronic kidney disease: a 6-month pilot scale trial in Canada. Curr Med Res Opin 2009;25:1919-30. [Crossref] [PubMed]
- Bammens B, Evenepoel P, Verbeke K, et al. Impairment of small intestinal protein assimilation in patients with end-stage renal disease: extending the malnutrition-inflammation-atherosclerosis concept. Am J Clin Nutr 2004;80:1536-43. [Crossref] [PubMed]
- Bammens B, Verbeke K, Vanrenterghem Y, et al. Evidence for impaired assimilation of protein in chronic renal failure. Kidney Int 2003;64:2196-203. [Crossref] [PubMed]
- Maraj M, Kuśnierz-Cabala B, Dumnicka P, et al. Malnutrition, Inflammation, Atherosclerosis Syndrome (MIA) and Diet Recommendations among End-Stage Renal Disease Patients Treated with Maintenance Hemodialysis. Nutrients 2018;10:69. [Crossref] [PubMed]
- Ridker PM, MacFadyen JG, Thuren T, et al. Effect of interleukin-1β inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet 2017;390:1833-42. [Crossref] [PubMed]
- Ridker PM, Everett BM, Thuren T, et al. Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. N Engl J Med 2017;377:1119-31. [Crossref] [PubMed]
- Syren ML, Turolo S, Marangoni F, et al. The polyunsaturated fatty acid balance in kidney health and disease: A review. Clin Nutr 2018;37:1829-39. [Crossref] [PubMed]
- Zeisel SH, Warrier M. Trimethylamine N-Oxide, the Microbiome, and Heart and Kidney Disease. Annu Rev Nutr 2017;37:157-81. [Crossref] [PubMed]
- Kaysen GA, Johansen KL, Chertow GM, et al. Associations of Trimethylamine N-Oxide With Nutritional and Inflammatory Biomarkers and Cardiovascular Outcomes in Patients New to Dialysis. J Ren Nutr 2015;25:351-6. [Crossref] [PubMed]
- Tang WH, Wang Z, Kennedy DJ, et al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res 2015;116:448-55. [Crossref] [PubMed]
- Ranganathan N, Ranganathan P, Friedman EA, et al. Pilot study of probiotic dietary supplementation for promoting healthy kidney function in patients with chronic kidney disease. Adv Ther 2010;27:634-47. [Crossref] [PubMed]
- Wang QJ, Shen YE, Wang X, et al. Concomitant memantine and Lactobacillus plantarum treatment attenuates cognitive impairments in APP/PS1 mice. Aging (Albany NY) 2020;12:628-49. [Crossref] [PubMed]
- Domenici L, Monti M, Bracchi C, et al. D-mannose: a promising support for acute urinary tract infections in women. A pilot study. Eur Rev Med Pharmacol Sci 2016;20:2920-5. [PubMed]
- Colletti A, Sangiorgio L, Martelli A, et al. Highly Active Cranberry's Polyphenolic Fraction: New Advances in Processing and Clinical Applications. Nutrients 2021;13:2546. [Crossref] [PubMed]
- Wang S, Xiao C, Liu C, et al. Identification of Biomarkers of Sepsis-Associated Acute Kidney Injury in Pediatric Patients Based on UPLC-QTOF/MS. Inflammation 2020;43:629-40. [Crossref] [PubMed]
- Jaisson S, Desmons A, Doué M, et al. Measurement of Homocitrulline, A Carbamylation-derived Product, in Serum and Tissues by LC-MS/MS. Curr Protoc Protein Sci 2018;92:e56. [Crossref] [PubMed]
- Sato T, Kawasaki Y, Maekawa M, et al. Accurate quantification of urinary metabolites for predictive models manifest clinicopathology of renal cell carcinoma. Cancer Sci 2020;111:2570-8. [Crossref] [PubMed]
- Kurella Tamura M, Chertow GM, Depner TA, et al. Metabolic Profiling of Impaired Cognitive Function in Patients Receiving Dialysis. J Am Soc Nephrol 2016;27:3780-7. [Crossref] [PubMed]
- Yi R, Tan F, Liao W, et al. Isolation and Identification of Lactobacillus plantarum HFY05 from Natural Fermented Yak Yogurt and Its Effect on Alcoholic Liver Injury in Mice. Microorganisms 2019;7:530. [Crossref] [PubMed]
- Chen YM, Wei L, Chiu YS, et al. Lactobacillus plantarum TWK10 Supplementation Improves Exercise Performance and Increases Muscle Mass in Mice. Nutrients 2016;8:205. [Crossref] [PubMed]
- Rice JC, Peng T, Spence JS, et al. Pyelonephritic Escherichia coli expressing P fimbriae decrease immune response of the mouse kidney. J Am Soc Nephrol 2005;16:3583-91. [Crossref] [PubMed]
- Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 2012;13:701-12. [Crossref] [PubMed]
- Dubourg G, Morand A, Mekhalif F, et al. Deciphering the Urinary Microbiota Repertoire by Culturomics Reveals Mostly Anaerobic Bacteria From the Gut. Front Microbiol 2020;11:513305. [Crossref] [PubMed]
- Staley C, Vaughn BP, Graiziger CT, et al. Gut-sparing treatment of urinary tract infection in patients at high risk of Clostridium difficile infection. J Antimicrob Chemother 2017;72:522-8. [Crossref] [PubMed]
- Murina F, Vicariotto F, Lubrano C. Efficacy of an orally administered combination of Lactobacillus paracasei LC11, cranberry and D-mannose for the prevention of uncomplicated, recurrent urinary tract infections in women. Urologia 2021;88:64-8. [Crossref] [PubMed]
- Milandri R, Bocchialini T, Maltagliati M, et al. Effects of D-Mannose, ElliroseTM and Lactobacillus Plantarum in treatment of urinary tract recurrent infections (rUTIs): A survey of urologists knowledge about its clinical application. Acta Biomed 2020;91:15-20. [PubMed]
- Mainini G, Passaro M, Schiattarella A, et al. Prevention and treatment of cystitis during menopause: efficacy of a nutraceutical containing D-mannose, inulin, cranberry, bearberry, Olea europaea, Orthosiphon and Lactobacillus acidophilus. Prz Menopauzalny 2020;19:130-4. [Crossref] [PubMed]
- Dahl WJ, Hung WL, Ford AL, et al. In older women, a high-protein diet including animal-sourced foods did not impact serum levels and urinary excretion of trimethylamine-N-oxide. Nutr Res 2020;78:72-81. [Crossref] [PubMed]
- Price TK, Dune T, Hilt EE, et al. The Clinical Urine Culture: Enhanced Techniques Improve Detection of Clinically Relevant Microorganisms. J Clin Microbiol 2016;54:1216-22. [Crossref] [PubMed]
- Hilt EE, McKinley K, Pearce MM, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol 2014;52:871-6. [Crossref] [PubMed]