
Chaetoglobosin E inhibits tumor growth and promotes the anti-tumor efficacy of cytotoxic drugs in esophageal squamous cell carcinoma by targeting PLK1
Introduction
Esophageal cancer is a common malignancy of the upper digestive tract, ranking eighth in terms of incidence among all tumors worldwide (1). Esophageal adenocarcinoma and esophageal squamous cell carcinoma (ESCC) are the most common subtypes of esophageal cancer, accounting for more than 95% of esophageal cancers (2). Esophageal adenocarcinoma is the prevailing subtype of esophageal cancer among Western populations, while ESCC is dominant among Asian populations (3). In China, the incidence rate of esophageal cancer is very high, accounting for more than 70% of cases globally (3). Moreover, 90% of patients in China have ESCC (3). Currently, the 5-year overall survival rate is about 10% and the 5-year survival rate after esophagectomy is only about 15–40% (4).
Chemotherapy, mainly including cisplatin, fluorouracil, and paclitaxel, plays an important role in the treatment of esophageal cancer, especially for advanced patients who cannot undergo surgery (5). However, these drugs have significant disadvantages, such as non-selective toxicity, considerable side effects, and poor patient compliance (6). With the development of immunotherapy, trastuzumab, ramucirumab, and pembrolizumab have been approved for the treatment of esophageal cancer; however, these agents need to be combined with chemotherapy to achieve good clinical effects; they are also expensive and have a limited potential patient pool (7). Therefore, there is a pressing need to develop novel anti-tumor agents.
Microbial metabolites are important sources of discovery of drug lead compounds (8). Chaetoglobosins are derived from fungal secondary metabolites and belong to the cytochalasan alkaloid class of drugs, which contain a 10-(indol-3-yl) group, a macrocyclic ring, and a perhydroisoindolone moiety (9). To date, around 100 chaetoglobosins and their analogues have been extracted and identified from a variety of fungi. They exert a broad range of biological activities, among which their anti-tumor activity has been studied most extensively (10). Previously, we reported the anti-tumor activities of a series of chaetoglobosins isolated from the metabolites of Chaetomium madrasense 375 (11-13). We found that chaetoglobosin E had a more potent anti-tumor effect against A549, HCC827, SW620, and MDA-MB-231 cells compared to those of chaetoglobosin B1, B2, B3, B, D, cytoglobosin A, and cisplatin (12). Moreover, several studies also have demonstrated that chaetoglobosin E, which is extracted from Chaetomium globosum, exerts anti-tumor activity against HeLa, HCT-116, and KB cells (14-16). However, these studies only performed cell proliferation analyses, and the underlying mechanism remains unclear. Our subsequent experiments demonstrated that chaetoglobosin E had the most potent inhibition on ESCC KYSE-30 cells compared with other cancer cells. Therefore, in this study, we explored the anti-proliferative activities and mechanisms of chaetoglobosin E (isolated from metabolites of Chaetomium madrasense 375) against KYSE-30 cells. Our study first elucidated that chaetoglobosin E induced pyroptosis via the activation of gasdermin E (GSDME) protein in ESCC cells. Importantly, we further found that polo-like kinase 1 (PLK1) might be its target and was involved in the regulation of pyroptosis, indicating that chaetoglobosin E induced cell pyroptosis by inhibiting PLK1. Thus, these findings suggested that chaetoglobosin E may be a novel lead compound to the treatment of ESCC patients by targeting PLK1, and elucidated for the first time that PLK1 was involved in a new pyroptosis mechanism. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5320/rc).
Methods
Reagents
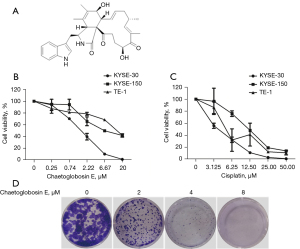
Chaetoglobosin E (purity ≥98%; molecular weight =528; Figure 1A) was isolated from the fungus Chaetomium madrasense 375 by the Comprehensive Utilization of Edible and Medicinal Plant Resources Engineering Technology Research Center. It was dissolved in dimethyl sulfoxide (DMSO) at a 20 mM concentration and stored at −20 ℃ until further use.
Cell cultures
Human esophageal cancer KYSE-30, KYSE-150, and TE-1 cells were purchased from the Cell Bank of the Chinese Academy of Sciences. They were cultured in the Roswell Park Memorial Institute (RPMI)-1640 medium containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin and incubated with 5% CO2 at 37 ℃.
Cell viability assay
Cells were seeded in 96-well plates (1×104 cells/well). After 24 h, the cells were treated with different concentrations of chaetoglobosin E for 48 h. Following 48 h of incubation, and the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) solution (Sigma-Aldrich, Darmstadt, Germany) was added into each well to achieve a final concentration of 0.5 mg/mL and then incubated for an additional 4 h. Subsequently, we carefully discarded the medium, and the formazan crystal was dissolved in 100 µL DMSO. A microplate spectrophotometer (Tecan, Switzerland) was used to measure the optical density.
Colony formation assay
Cells were seeded in 24-well plates (500 cells/well). After 24 h, the cells were treated with different concentrations of chaetoglobosin E. A medium containing the appropriate concentrations of chaetoglobosin E was changed every 2 days. After 10 days, the colonies were fixed and stained with 1% crystal violet.
Cell cycle assay
KYSE-30 cells were seeded in six-well plates (3×105 cells/well). After 24 h, cells were treated with different concentrations of chaetoglobosin E for 48 h, and then collected and washed with phosphate-buffered saline (PBS). Subsequently, the cells were fixed with 70% ethanol at −20 ℃ overnight. The following day, cells were treated with a cell cycle detection kit (Keygen, China) and analyzed using FACSCalibur flow cytometry (BD, USA).
RNA-sequencing assay
KYSE-30 cells were seeded in six-well plates (3×105 cells/well). After 24 h, the cells were treated with 8 µM chaetoglobosin E for 48 h. Thereafter, the cell supernatant was discarded and cells were washed with PBS. Subsequently, the cells were lysed using Trizol (Sigma-Aldrich, Darmstadt, Germany), and the lysates were collected for an RNA-sequencing (RNA-seq) assay.
Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
KYSE-30 cells were seeded in six-well plates (3×105 cells/well). After 24 h, the cells were treated with different concentrations of chaetoglobosin E for 48 h, and the total RNA from the cultured cells was then prepared using the RNeasy Mini Kit (Qiagen, Germany). A 200 ng sample of total RNA was subjected to RT-PCR using the GoScript™ Reverse Transcription System kit (Takara, Japan) and TB Green™ Premix Ex Taq™ II (Tli RNaseH Plus) kit (Takara, Japan). The amplification process was performed in 40 cycles at 95 ℃ for 30 s, 95 ℃ for 5 s, and 60 ℃ for 31 s.
The primer sequences used in this study were as follows: PLK1 forward: 5'-GTTCCCATCCCAACTCCTTGA-3', reverse: 5'-TGCTCGCTCATGTAATTGCG-3'; and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward: 5'-CGGATTTGGTCGTATTGGG-3', reverse: 5'-CTGGAAGATGGTGATGGGATT-3'. The levels of RNA were analyzed using the 2-∆∆CT method and all quantifications were normalized to the level of the internal control gene, GAPDH.
Western Blot assay
KYSE-30 cells were seeded in six-well plates (3×105 cells/well). After 24 h, the cells were treated with different concentrations of chaetoglobosin E for 48 h, and then collected and washed with PBS. The cellular proteins were extracted using a protein extraction kit (Keygen, China). The proteins were separated in 10% sodiumdodecyl sulfate-poly acrylamide gel electrophoresis (SDS-PAGE) gels according to their different molecular weights, and the separated proteins were transferred to a polyvinylidene fluoride (PVDF) membrane by wet rotation. The PVDF membranes were then blocked with 5% skim milk for 1 h and incubated with primary antibodies to ERK, p-ERK, p-MEK, Akt, p-Akt (Cell Signaling Technology, America), PLK1 (Abcam, UK), MEK, EGFR, p-EGFR, GSDME, Bcl-2, Bax, beclin1, LC3, p21, cyclinB, CDC2, p-CDC2, E-cadherin, vimentin, and Actin (Abways, China) at 4 ℃ overnight. The following day, the PVDF membranes were washed with tris buffered saline-Tween-20 (TBST) and incubated with the appropriate secondary antibody for 1 h. The protein bands were imaged using the BLT GelView 6000Plus (Guangzhou, China).
Overexpression and silencing of PLK1
KYSE-30 cells were seeded in six-well plates (3×105 cells/well). After 24 h, the cells were transfected with a pcDNA3.1(+)-PLK1 plasmid (2 µg), an empty pcDNA3.1(+) plasmid (2 µg), an siRNA for PLK1 (100 pmol), or a negative control siRNA-A (100 pmol, Santa Cruz, America) using lip2000 (Invitrogen, America). After 6 h, the cells were washed and a new medium was added for 48 h, and the total proteins were extracted thereafter. The expressions of the PLK1 and GSDME proteins were analyzed by western blot.
Correlation assay of anti-tumor activity of chaetoglobosin E and PLK1 expression
KYSE-30 cells were seeded in six-well plates (3×105 cells/well). After 24 h, the cells were transfected with a pcDNA3.1(+)-PLK1 plasmid (0.5, 1 µg) or an empty pcDNA3.1(+) plasmid (0.5, 1 µg). After 6 h, the cell supernatant was discarded and the cells were added to the new medium with or without 4 µM chaetoglobosin E. After 48 h, the total RNA and total protein were extracted. The RNA and protein expressions of PLK1 were analyzed by real-time qRT-PCR and western blot, respectively.
Synergistic effects against tumor activity of chaetoglobosin E with cytotoxic agents
The KYSE-30 cells were treated with different concentrations of chaetoglobosin E or cytotoxic agents [cisplatin, 5-fluorouracil (5-Fu)] alone, or co-treated with chaetoglobosin E and cytotoxic agents (cisplatin, 5-Fu). After 48 h of incubation, an MTT solution (Sigma-Aldrich) was added into each well to achieve a final concentration of 0.5 mg/mL and then incubated for an additional 4 h. Subsequently, we carefully discarded the medium, and the formazan crystal was dissolved in 100 µL DMSO. A microplate spectrophotometer (Tecan) was used to measure the optical density. The combination index (CI) was calculated by the Chou-Talalay method using CompuSyn by (version 1.0); CI >1 denoted antagonism, CI =1 indicated addition, and CI <1 suggested synergy between the two drugs used.
Statistical analysis
Data were presented as the mean ± standard deviation (SD). Statistical analyses were carried out using GraphPad Prism 5.0 and SPSS software version 23.0. Statistical evaluation was performed using one-way analysis of variance (ANOVA) and Tukey’s test, and P<0.05 was considered statistically significant.
Results
Chaetoglobosin E inhibited cell viability and colony formation
The effect of chaetoglobosin E on the viability of esophageal cancer cells was assessed. Chaetoglobosin E exhibited potent cytotoxic activity against three esophageal cancer cells, including KYSE-30, KYSE-150, and TE-1 cells, in a dose-dependent manner, especially for ESCC KYSE-30 cells (Figure 1B). Compared with other cancer cells reported in the literature, chaetoglobosin E also exhibited the most potent inhibition on KYSE-30 cells (50% inhibitive concentration (IC50) value of 2.57 µmol/L), indicating that chaetoglobosin E can be used as a new lead compound against ESCC (17-19). Therefore, KYSE-30 cells were used for subsequent experiments. Moreover, cisplatin, as a positive control, also showed a significant cytotoxicity effect in these cancer cells (Figure 1C).
Colony formation ability was assessed to explore the long-term impact of chaetoglobosin E on cancer cell growth. The colony-forming ability of KYSE-30 cells was significantly suppressed by chaetoglobosin E in a dose-dependent manner. These findings indicated that the treatment of KYSE-30 cells with chaetoglobosin E inhibited cell proliferation and colony formation (Figure 1D).
Chaetoglobosin E arrested cell cycle progression at the G2/M phase
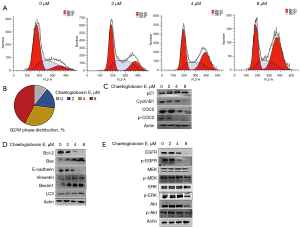
To explore the anti-tumor mechanism of chaetoglobosin E, we analyzed its effect on the cell cycle by flow cytometry. Chaetoglobosin E treatment markedly arrested cell cycle progression at the G2/M phase in a dose-dependent manner (Figure 2A,2B). Consistent with the flow cytometry analysis, chaetoglobosin E treatment decreased the G2/M phase regulatory protein expressions of cyclinB1, CDC2, and p-CDC2, and increased the protein expression of p21 (Figure 2C). These results suggested that chaetoglobosin E might inhibit cell growth by inducing G2/M phase arrest, followed by the down-regulation of cyclinB1, CDC2, and p-CDC2 and the up-regulation of p21.
Chaetoglobosin E induced cell apoptosis and autophagy, and inhibited invasion and metastasis
We then further explored the effect of chaetoglobosin E on cell apoptosis, autophagy, invasion, and metastasis by western blot. Chaetoglobosin E treatment decreased the anti-apoptotic protein expression of Bcl-2, increased the apoptotic expression of Bax, increased the autophagy protein expressions of beclin1 and LC3, decreased the invasion and metastasis protein expression of E-cadherin, and increased the expression of vimentin (Figure 2D).
Chaetoglobosin E inhibited the EGFR/MEK/ERK and Akt signaling pathways
It is well established that the EGFR/MEK/ERK and Akt signaling pathways are closely related to multiple processes of cell growth and play important roles in the development of tumorigenesis (17). Thus, we explored whether these pathways participated in the chaetoglobosin E-mediated anti-tumor activity. The treatment of chaetoglobosin E decreased the protein expressions of p-EGFR, p-MEK, p-ERK, and p-Akt (Figure 2E). Our findings suggested that chaetoglobosin E might induce the cell G2/M phase arrest, apoptosis, and autophagy, and inhibit invasion and metastasis by inhibiting the EGFR/MEK/ERK and Akt signaling pathways.
Chaetoglobosin E exerted anti-tumor effect by targeting PLK1
Subsequently, an RNA-seq assay was used to identify the specific target of chaetoglobosin E. The results showed that the treatment of KYSE-30 cells with chaetoglobosin E significantly decreased PLK1 expression, which was closely related to the cell G2/M phase, apoptosis, autophagy, invasion, and metastasis. According to the Gene Expression Profiling Interactive Analysis (GEPIA) database, the expression of PLK1 in esophageal cancer tissue is notably higher than that in para-carcinoma tissue, indicating that it is involved with the progression of esophageal cancer (Figure 3A). Therefore, we wondered whether this protein participated in the chaetoglobosin E-mediated anti-tumor activity.
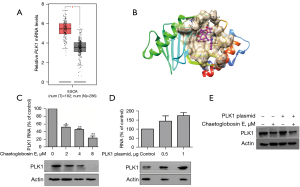
Large loop prediction software was used to generate 76 conformations of chaetoglobosin E and analyze the interaction of these conformations with the PLK1 protein. The results suggested that chaetoglobosin E had a good interaction with the PLK1 protein (Figure 3B). Also, the impact of chaetoglobosin E on PLK1 was further verified by real-time qRT-PCR and western blot. The real-time qRT-PCR results showed that chaetoglobosin E decreased PLK1 gene expression in a dose-dependent manner (Figure 3C). Consistent with this, the western blot results also demonstrated that chaetoglobosin E decreased the protein expression of PLK1 in a dose-dependent manner (Figure 3C). Subsequently, we used a PLK1-specific plasmid to evaluate the effect of chaetoglobosin E on PLK1. Cells were transfected with the PLK1-specific plasmid and concurrently treated with or without chaetoglobosin E. The treatment of chaetoglobosin E reversed the PLK1 plasmid-induced up-regulation of the PLK1 protein level, suggesting that chaetoglobosin E exerted its anti-tumor effect by targeting PLK1 (Figure 3D,3E).
Chaetoglobosin E induced cell pyroptosis by inhibiting PLK1
Previous reports have demonstrated that there is a transition between cell apoptosis and pyroptosis in cancer (12,13). The present study indicated that chaetoglobosin E induced cell apoptosis; thus, we further explored whether chaetoglobosin E affected pyroptosis. Chaetoglobosin E treatment can activate the pyroptosis biomarker GSDME in a dose-dependent manner, suggesting that chaetoglobosin E induced cell pyroptosis (Figure 4A).
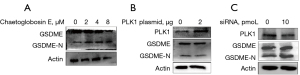
Subsequently, we also analyzed the impact of PLK1 on pyroptosis. KYSE-30 cells were transfected with a PLK1 plasmid or PLK1-specific siRNA. The up-regulation of PLK1 inhibited the activation of GSDME, while the down-regulation of PLK1 promoted the activation of GSDME, suggesting that PLK1 was involved in the regulation of pyroptosis (Figure 4B,4C). Taken together, our findings suggested that chaetoglobosin E induced cell pyroptosis by inhibiting PLK1.
The synergistic anti-proliferative effect of chaetoglobosin E and cisplatin/5-Fu
Considering the anti-tumor mechanism of chaetoglobosin E, we speculated that it might be a good anti-tumor drug, in combination with cytotoxic agents. To confirm this, KYSE-30 cells were treated with chaetoglobosin E plus cisplatin or 5-Fu. The results showed that chaetoglobosin E inhibited the proliferation of KYSE-30 cells synergistically when used together with cisplatin or 5-Fu, with a CI <1.0 at the 50% effective concentration (EC50) concentration and a dose-reduction index (DRI) >1.0 (Table 1).
Table 1
| Drugs | EC50 (μM) | CIa | DRIb | ||
|---|---|---|---|---|---|
| Chaetoglobosin E | Cisplatin | 5-Fu | |||
| Chaetoglobosin E | 3.68 | ||||
| Cisplatin | 3.55 | ||||
| 5-Fu | 30.56 | ||||
| Chaetoglobosin E+ cisplatin | 1.40+1.40 | 0.77 | 2.64 | 2.54 | |
| Chaetoglobosin E + 5-Fu | 0.89+8.88 | 0.50 | 4.88 | 3.44 | |
a, CI were evaluated by calculating by the Chou-Talalay method using CompuSyn version 1.0. b, folds of dose reduction allowed for each drug caused by synergism at EC50 effect level. 5-Fu, 5-fluorouracil; EC50, 50% effective concentration; CI, combination index; DRI, dose reduction index.
Discussion
Currently, the overall effect of esophageal cancer therapy is still not satisfactory, and thus, there is an urgent need to develop novel anti-tumor agents. Chaetoglobosins have attracted considerable attention due to their structural diversity and wide-ranging bioactivities (10). In this study, chaetoglobosin E, which is isolated from the secondary metabolites of Chaetomium madrasense 375, exhibited potent inhibition on three esophageal cancer cells including KYSE-30, KYSE-150, and TE-1 cells, in a dose-dependent manner, especially on the ESCC KYSE-30 cells. Previously, we reported that chaetoglobosin E exerts anti-tumor activity on several cancer cells, including lung cancer A549 and HCC827 cells, colon cancer SW620 cells, and breast cancer MDA-MB-621 cells (12). Moreover, research by other groups also demonstrated that chaetoglobosin E exhibits cytotoxicity against HeLa, HCT116, and KB cells (14-16). In the present study, we found that, compared with other cancer cells, chaetoglobosin E exhibited the most potent inhibition on ESCC KYSE-30 cells (IC50 value of 2.57 µmol/L), indicating that chaetoglobosin E can be used as a new lead compound against esophageal cancer. However, these experiments were limited to in vitro cell proliferation analyses, and the anti-tumor mechanism remained unclear.
Several previous studies have confirmed that the cell cycle is a crucial factor affecting tumor growth (20,21). Our results showed that chaetoglobosin E induced G2/M phase arrest of KYSE-30 cells, followed by the down-regulation of cyclinB1, CDC2, and p-CDC2, and the up-regulation of p21. The cell cycle and apoptosis are closely related, with several common genes (22). Subsequently, we found that chaetoglobosin E treatment decreased the anti-apoptotic protein expression of Bcl-2 and increased the apoptotic expression of Bax. Apoptosis and autophagy are two major types of programmed cell death. Some studies have shown that apoptosis may decrease autophagy (23,24). For example, Luo et al. found that the key autophagy protein Beclin1 can be negatively regulated by Bcl-2 and Bcl-xL, indicating the suppression of autophagy by apoptosis (23). Another study by Oral et al. also showed that autophagy related protein 3 (Atg3) can be cleaved by caspase-8 to inhibit autophagy (24). However, some studies have also found that autophagy may help to induce apoptosis (25,26).
We then explored the effect of chaetoglobosin E on autophagy. Our results indicated that chaetoglobosin E treatment increased the expression of autophagy proteins beclin1 and LC3. Moreover, we also found that chaetoglobosin E treatment decreased the expression of the invasion and metastasis protein E-cadherin and increased the expression of vimentin. From the above results, this study confirmed for the first time that chaetoglobosin E induced ESCC cell arrest at the G2/M phase, cell apoptosis, and autophagy, and inhibited cell invasion and metastasis.
The RNA-seq assay is an important measurement tool to quantify messenger RNAs (mRNAs) for gene expression (27). We used an RNA-seq assay to explore the specific target of chaetoglobosin E, and our results showed that the treatment of KYSE-30 cells with chaetoglobosin E significantly decreased the expression of PLK1 in a dose-dependent manner. Mammalian PLK family members include PLK1, PLK2, PLK3, PLK4, and PLK5, which are serine/threonine protein kinases (28). PLK1 is highly conserved from yeast to humans (29) and is involved in various tumor progression processes via its close relationship with cell G2/M phase, apoptosis, autophagy, invasion, and metastasis (30-33). According to the GEPIA database, the expression of PLK1 in esophageal cancer tissue is significantly higher than that in para-carcinoma tissue, highlighting its involvement with esophageal cancer progression. Moreover, large loop prediction software analysis showed that chaetoglobosin E had a good interaction with the PLK1 protein. Our other experiments also verified that chaetoglobosin E decreased the gene and protein expressions of PLK1 in a dose-dependent manner. Subsequently, we used a PLK1-specific plasmid to evaluate the effect of chaetoglobosin E on PLK1 and found that chaetoglobosin E reversed PLK1 plasmid-induced up-regulation of the PLK1 protein level. These results suggested that chaetoglobosin E exerted its anti-tumor effect by targeting PLK1.
Pyroptosis is a gasdermin (GSDM)-dependent regulated mode of cell death and plays a major role in the occurrence and development of various diseases, including tumors, infections, nervous system diseases, and atherosclerosis (34-36). Previous studies have demonstrated that there is a close relationship between pyroptosis, apoptosis, and autophagy (18,19,37). For example, Meng et al. found that sesamin, a lignan compound in plants, promoted apoptosis and pyroptosis via the activation of autophagy to enhance the anti-tumor action on murine T-cell lymphoma (35). Furthermore, reports have found that chemotherapeutics induce pyroptosis through the BAK/BAX-caspase-3-GSDME pathway, and caspase-3 causes a transition between apoptosis and pyroptosis (18,19). Therefore, we speculated that chaetoglobosin E may affect cell pyroptosis.
Our results also demonstrated that chaetoglobosin E induced pyroptosis via activation of the GSDME protein, indicating that chaetoglobosin E might also inhibit the proliferation of KYSE-30 cells by promoting pyroptosis. So, we further assessed whether PLK1 participate in cell pyroptosis. Further experiments demonstrated that the high expression of PLK1 inactivated the GSDME protein, while the knockdown of PLK1 expression activated this protein. Therefore, chaetoglobosin E may induce cell pyroptosis by inhibiting PLK1.
Our results demonstrated that chaetoglobosin E significantly inhibited the proliferation of KYSE-30 cells. Mechanistically, this is the first report showing that chaetoglobosin E might inhibit PLK1 to induce the G2/M phase arrest of KYSE-30 cells as well as apoptosis, autophagy, and pyroptosis and inhibit cell invasion and metastasis.
Conclusions
This study demonstrated that chaetoglobosin E may be a novel lead compound for the treatment of ESCC patients by inhibiting PLK1, and elucidated for the first time that PLK1 is involved in a new mechanism of pyroptosis. This study provided a good lead compound, and it will be interesting to use chaetoglobosin E to structurally modify to develop more active drugs for ESCC treatment. Moreover, it revealed a new molecular mechanism of PLK1 for ESCC treatment. Thus, these findings will help to promote the further treatment and improve the prognosis of ESCC. However, the detailed mechanism of chaetoglobosin E inhibition of PLK1 remains unclear and needs to explored in future research.
Acknowledgments
Funding: This work was supported by the Tackling-plan Project of Henan Department of Science and Technology (Nos. 212102310325, 212102310243, 202102310489, and 192102310053), the Tackling-plan Project of Henan Medical Science and Technology (No. LHGJ20200164), and the Innovation and Entrepreneurship Training Program for College students in Henan Province (No. 202111834037).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5320/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5320/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5320/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Reichenbach ZW, Murray MG, Saxena R, et al. Clinical and translational advances in esophageal squamous cell carcinoma. Adv Cancer Res 2019;144:95-135. [Crossref] [PubMed]
- Uhlenhopp DJ, Then EO, Sunkara T, et al. Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin J Gastroenterol 2020;13:1010-21. [Crossref] [PubMed]
- Wang Q, Liu H, Liu Z, et al. Circ-SLC7A5, a potential prognostic circulating biomarker for detection of ESCC. Cancer Genet 2020;240:33-9. [Crossref] [PubMed]
- Huang FL, Yu SJ. Esophageal cancer: Risk factors, genetic association, and treatment. Asian J Surg 2018;41:210-5. [Crossref] [PubMed]
- Ikeda G, Yamamoto S, Kato K. The safety of current treatment options for advanced esophageal cancer after first-line chemotherapy. Expert Opin Drug Saf 2022;21:55-65. [Crossref] [PubMed]
- Ku GY. Systemic therapy for esophageal cancer: chemotherapy. Chin Clin Oncol 2017;6:49. [Crossref] [PubMed]
- Lu Y, Xu M, Guan L, et al. PD-1 Inhibitor Plus Chemotherapy Versus Chemotherapy as First-line Treatment for Advanced Esophageal Cancer: A Systematic Review and Meta-Analysis. J Immunother 2022;45:243-53. [Crossref] [PubMed]
- Greco C, Keller NP, Rokas A. Unearthing fungal chemodiversity and prospects for drug discovery. Curr Opin Microbiol 2019;51:22-9. [Crossref] [PubMed]
- Huang S, Chen H, Li W, et al. Bioactive Chaetoglobosins from the Mangrove Endophytic Fungus Penicillium chrysogenum. Mar Drugs 2016;14:172. [Crossref] [PubMed]
- Chen J, Zhang W, Guo Q, et al. Bioactivities and Future Perspectives of Chaetoglobosins. Evid Based Complement Alternat Med 2020;2020:8574084. [PubMed]
- Chen J, Guo Q, Zhang J, et al. Chaetoglobosin G inhibits proliferation, autophagy and cell cycle of lung cancer cells through EGFR/MEK/ERK signaling pathway. Pharmazie 2020;75:642-5. [PubMed]
- Guo Q, Chen J, Ren Y, et al. Hydrazine-Containing Heterocycle Cytochalasan Derivatives From Hydrazinolysis of Extracts of a Desert Soil-Derived Fungus Chaetomium madrasense 375. Front Chem 2021;9:620589. [Crossref] [PubMed]
- Guo QF, Yin ZH, Zhang JJ, et al. Chaetomadrasins A and B, Two New Cytotoxic Cytochalasans from Desert Soil-Derived Fungus Chaetomium madrasense 375. Molecules 2019;24:3240. [Crossref] [PubMed]
- Shen L, Zhu L, Wei ZQ, et al. Chemical constituents from endophyte Chaetomium globosum in Imperata cylindrical. Zhongguo Zhong Yao Za Zhi 2015;40:4645-9. [PubMed]
- Zheng QC, Kong MZ, Zhao Q, et al. Chaetoglobosin Y, a new cytochalasan from Chaetomium globosum. Fitoterapia 2014;93:126-31. [Crossref] [PubMed]
- Ding G, Song YC, Chen JR, et al. Chaetoglobosin U, a cytochalasan alkaloid from endophytic Chaetomium globosum IFB-E019. J Nat Prod 2006;69:302-4. [Crossref] [PubMed]
- Levantini E, Maroni G, Del Re M, et al. EGFR signaling pathway as therapeutic target in human cancers. Semin Cancer Biol 2022; Epub ahead of print. [Crossref] [PubMed]
- Hu L, Chen M, Chen X, et al. Chemotherapy-induced pyroptosis is mediated by BAK/BAX-caspase-3-GSDME pathway and inhibited by 2-bromopalmitate. Cell Death Dis 2020;11:281. [Crossref] [PubMed]
- Bertheloot D, Latz E, Franklin BS. Necroptosis, pyroptosis and apoptosis: an intricate game of cell death. Cell Mol Immunol 2021;18:1106-21. [Crossref] [PubMed]
- Li S, Wang L, Wang Y, et al. The synthetic lethality of targeting cell cycle checkpoints and PARPs in cancer treatment. J Hematol Oncol 2022;15:147. [Crossref] [PubMed]
- Vlatkovic T, Veldwijk MR, Giordano FA, et al. Targeting cell cycle checkpoint kinases to overcome intrinsic radioresistance in brain tumor cells. Cancers (Basel) 2022;14:701. [Crossref] [PubMed]
- Vermeulen K, Berneman ZN, Van Bockstaele DR. Cell cycle and apoptosis. Cell Prolif 2003;36:165-75. [Crossref] [PubMed]
- Luo S, Rubinsztein DC. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ 2010;17:268-77. [Crossref] [PubMed]
- Oral O, Oz-Arslan D, Itah Z, et al. Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis 2012;17:810-20. [Crossref] [PubMed]
- Jiang F, Bian G, Li J. ASPP2 promotes cell apoptosis in cervical cancer through inhibiting autophagy. Exp Ther Med 2022;24:726. [Crossref] [PubMed]
- Huang J, Ye Y, Xiao Y, et al. Geniposide ameliorates glucocorticoid-induced osteoblast apoptosis by activating autophagy. Biomed Pharmacother 2022;155:113829. [Crossref] [PubMed]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57-63. [Crossref] [PubMed]
- de Cárcer G, Manning G, Malumbres M. From Plk1 to Plk5: functional evolution of polo-like kinases. Cell Cycle 2011;10:2255-62. [Crossref] [PubMed]
- Bahassi el M. Polo-like kinases and DNA damage checkpoint: beyond the traditional mitotic functions. Exp Biol Med (Maywood) 2011;236:648-57. [Crossref] [PubMed]
- Barr FA, Silljé HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol 2004;5:429-40. [Crossref] [PubMed]
- Nieto-Jimenez C, Galan-Moya EM, Corrales-Sanchez V, et al. Inhibition of the mitotic kinase PLK1 overcomes therapeutic resistance to BET inhibitors in triple negative breast cancer. Cancer Lett 2020;491:50-9. [Crossref] [PubMed]
- Ruf S, Heberle AM, Langelaar-Makkinje M, et al. PLK1 (polo like kinase 1) inhibits MTOR complex 1 and promotes autophagy. Autophagy 2017;13:486-505. [Crossref] [PubMed]
- Fu Z, Wen D. The Emerging Role of Polo-Like Kinase 1 in Epithelial-Mesenchymal Transition and Tumor Metastasis. Cancers (Basel) 2017;9:131. [Crossref] [PubMed]
- Yang F, Bettadapura SN, Smeltzer MS, et al. Pyroptosis and pyroptosis-inducing cancer drugs. Acta Pharmacol Sin 2022;43:2462-73. [Crossref] [PubMed]
- Meng Z, Liu H, Zhang J, et al. Sesamin promotes apoptosis and pyroptosis via autophagy to enhance antitumour effects on murine T-cell lymphoma. J Pharmacol Sci 2021;147:260-70. [Crossref] [PubMed]
- Hu Y, Wang B, Li S, et al. Pyroptosis, and its Role in Central Nervous System Disease. J Mol Biol 2022;434:167379. [Crossref] [PubMed]
- Meng Z, Liu H, Zhang J, et al. Sesamin promotes apoptosis and pyroptosis via autophagy to enhance antitumour effects on murine T-cell lymphoma. J Pharmacol Sci 2021;147:260-70. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


