
MYC gene associated polymorphisms and Wilms tumor risk in Chinese children: a four-center case-control study
Introduction
Wilms tumor (WT), also known as nephroblastoma, is the most prevalent pediatric renal cancer. It accounts for 6–7% of all childhood malignancies occurring in children younger than 15 years (1). The incidence of WT varies from one ethnic group to another, with the highest rate found in black African and the lowest rate in Asians (1). In the United States, the morbidity of WT is about 1 in 8,000 children, accompanying with 400–650 new cases yearly (2), while 1 per 10,000 children is diagnosed in Western descent (3). In China, the incidence rate of WT is approximately 3.3 per million, lower than that in Western countries (4). The various incidences of WT among different ethnicities reflect the implication of genetic backgrounds in the pathogenesis of this disease. Now, WT is believed to derive from embryonal nephric mesenchymal pluripotent precursor cells (5). With unknown factors, these cells fail to differentiate to nephrons and form various lesions (nephrogenic rests) instead, leading to the development of WT eventually (6). Great progress has been made in the treatment of WT. The survival rate has reached over 90% in patients with regional WT (7), and over 75% in those with metastatic disease (8). Despite the encouraging clinical outcomes, about 25% of survivors suffer chronic disorders. They are often high-risk patients with poor histologic and molecular characteristics, bilateral disease and relapse (9).
WT is a genetically heterogeneous and intricate disorder. The majority cases are sporadic, and only 1–2% are familial (10,11). To data, many WT susceptibility genes and epigenetic alterations have been reported (12). Wilms tumor gene 1 (WT1) at 11p13 was a tumor suppressor, the mutation in which was first identified in WT. It encodes a transcription factor important in multiple phases of normal kidney. And the somatic mutations of WT1 are seen in 10% to 20% of sporadic WT (10). WT1 mutation also frequently accompany with canonical Wnt activation, due to the activating mutation of β-catenin (CTNNB1) (13). WTX encodes a protein facilitating β-catenin degradation. Its inactivating mutations may occur in 15% to 20% WT patients (14). It was reported that the mutations of WT1, CTNNB1 and WTX are implicated in nearly 1/3 WT cases (15). Other genetic loci implicated in WT infrequently are the familial predisposition loci FWT1 at 17q12-q21 and FWT2 at 19q13.4 (16). And the TRIP13, which encodes a highly conserved AAA + ATPase that contributes to homolog pairing, synapsis, and recombination during meiosis. The biallelic loss-of-function mutations of TRIP13 are prone to chromosome segregation dysfunction and confer a high risk of WT (17). Besides, the Q177R mutation of the transcription factors SIX1/2 shifts DNA binding specificity, which may induce subtle changes in the gene regulatory capacity of SIX1/2, and thereby change the expression profile of downstream target gene. The mutations in the microRNA processing genes DROSHA/DGCR8 affect the miRNA processing and maturing, then alter miRNA expression patterns, such as all members of the miR-200 family and Let-7a. All of these mutations were reported to be involved in the development and progression of WT (18,19). Recent research reported that germline mutations of CHEK2 may reduce its kinase activity and affect RNA splicing, therefore involving in the progress of WT. The MYCN germline mutations that lead to aberrant activation, the deletions of DIS3L2, the HACE1 germline mutations that lead to promoter methylation and the germline mutations of CDKN2A and CDKN2B that lead to their homozygous deletion will all play a role in the WT development (20). Furthermore, SNPs in several genes including BARD1 (21), LIN28 (22), miR-423 (23) and ERCC2 (24) have been found to predispose to WT.
MYC is located at chromosome 8 (8q24.21). It encodes a nuclear phosphoprotein transcription factor that plays key roles in the regulation of cellular proliferation, growth, apoptosis, metabolic transformation and oncogenesis (25). MYC family of proto-oncogenes is one of the most studied oncogenes (26). MYC is activated in approximately 70% of all human cancers and regulates about 10% of human genes involved in cellular malignant properties, such as promoting of cell proliferation and blocking cellular differentiation (27). Deregulation of MYC has been discovered in a great variety of human cancer, such as prostate cancer (28), and breast cancer (29). Single-nucleotide polymorphisms (SNPs) in the MYC are likely to lead to alterations in the structure and function of the protein (30), as well as the expression levels of MYC, thus contribute to cancer susceptibility (31). Several studies have reported numerous SNPs on chromosome 8q24 which contribute to cancer susceptibility of colorectal (32), prostate (33), breast cancer (34). However, as far as we know, there no reports about the associations between SNPs of MYC and WT susceptibility. Given this, we aimed to explore the association of two potentially functional SNPs (rs4645943 C > T, rs2070583 A > G) of MYC with WT risk in Chinese population through a four-center case-control study.
Methods
Study subjects
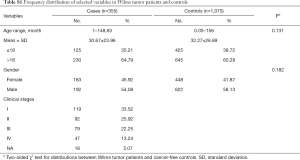
In this present study, a total of 355 cases with WT and 1,070 healthy controls were included (35,36). The 355 cases were collected from four hospitals (Guangzhou Women and Children’s Medical Center, The First Affiliated Hospital of Zhengzhou University, The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University, and Second Affiliated Hospital of Xi’an Jiao Tong University), and the 1,070 control subjects were enrolled randomly from the same areas at the same time (35,36). The demographic characteristics of all participants are displayed in Table S1. Written informed consent was obtained for each participant from their parents or guardians. This study got approved by each institutional review board of all participating hospitals. The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Full table
Polymorphism selection and genotyping
From the dbSNP database (http://www.ncbi.nlm.nih.gov/) and SNPinfo (http://snpinfo.niehs.nih.gov/), two potentially functional polymorphisms (rs4645943 C > T, and rs2070583 A > G) of MYC gene were chosen base on the following selection criteria: (I) position located at exons, splice sites, 5' near gene, 5' untranslated regions (UTR), 3' near gene, 3' UTR; (II) the minor allele frequency (MAF) should be ≥5% in Chinese Han population; (III) potentially functional SNPs are identified by SNPinfo software (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm); (IV) selected SNPs were in low linkage disequilibrium (LD) with one another using an R2 threshold lower than 0.8; (V) not explored in the published genome-wide association studies (GWASs) of WT (37). The rs4645943 C > T is a variant located in transcription start site upstream 2 Kb of MYC gene and the rs2070583 A > G, a SNP located in the MYC gene 3' UTR region, which may associate with the expression level of the MYC gene. For genotyping, the genomic DNA was extracted from peripheral blood leukocytes of all participants by the TIANamp Blood DNA Kit (TianGen Biotech, Beijing, China). The DNA samples were added to 96-well plates and diluted to 5 ng/µL. Genotyping for the SNPs was carried out in the 384-well format by the Taqman method. And ten percent of the samples were chosen randomly and re-genotyped. The concordance of the two sets of genotyping results was 100%.
Statistical analysis
χ2 test was applied to assess the differences in demographic characteristics and the frequency distributions of genotypes between WT cases and healthy controls. And the deviation from Hardy-Weinberg equilibrium (HWE) was evaluated by the goodness-of-fit χ2 test in the control subjects. The assessment of relevance between MYC gene associated polymorphisms and WT susceptibility was performed by calculating odds ratios (ORs) and 95% confidence intervals (CIs). Furthermore, by unconditional multivariate logistic regression analysis, adjusted ORs and corresponding 95% CIs which adjusted for age and gender were calculated. Stratified analyses were conducted based on the age, gender and clinical stages. All statistical tests were two-sided and analyzed using SAS software (version 9.4; SAS Institute, Cary, NC, USA). And when the P values <0.05 were considered as statistically significant.
Results
Relevance between MYC gene associated polymorphisms and WT risk
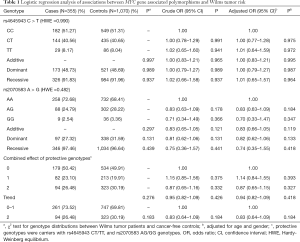
In this current case-control study, 355 cases and 1,070 controls were successfully genotyped. The genotype frequencies of two MYC gene associated polymorphisms are shown in Table 1, which were in accordance with HWE amongst the control subjects (P=0.990 for rs4645943 C > T; and P=0.482 for rs2070583 A > G). In single genotype analysis, no significant association was found between the two selected polymorphisms and the WT risk. And the same result was found in the combined analysis.
Full table
Stratification analysis of MYC gene associated polymorphisms with WT susceptibility
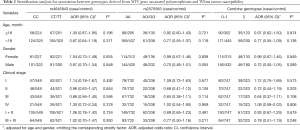
To explore whether the selected MYC gene associated polymorphisms affect WT susceptibility among different subgroups, stratified analyses were performed according to the age, gender and clinical stages (Table 2). However, no significant association was identified between the studied polymorphisms and the WT susceptibility in any subgroup.
Full table
Discussion
We conducted the present hospital-based study comprising 355 WT cases and 1,070 control subjects to investigate the relationship between two MYC SNPs and the WT risk. However, neither of the two selected SNPs was associated with WT risk. To the best of our knowledge, this is the first research exploring the association between polymorphisms in the MYC gene and WT risk.
As a proto-oncogene, MYC encodes a transcription factor that regulates the expression of approximately 15% of human genes directly or indirectly (38). It can interact with its partner protein MAX and bind to E-box DNA elements located in regulatory regions of target genes, then activate gene expression (39). Moreover, as MYC participates in the biogenesis of multiple components of the ribosome, it can regulate gene expression indirectly at the translational level (40). Importantly, MYC is a multifunctional protein, it acts as a master regulator that commands cellular proliferation, differentiation, metabolism, migration/invasion, apoptosis, microenvironment remodeling, angiogenesis, and immune responses by involving in multiple signaling pathways (41-43).
Deregulation of MYC has been repeatedly reported in a wide spectrum of human cancer, including breast cancer (44), and lung cancer (45). The deregulation of MYC leads to the aberrant expression of its downstream target genes, then causes the changes in the biological behaviors and functions, and promotes oncogenesis eventually. Although the major causes of oncogene deregulation are DNA amplification and gene mutation, SNPs may also cause an abnormality of gene structure and gene expression, then lead to functional change. There are plenty of studies reporting that SNPs in the MYC gene result in the dysregulation of MYC, and then modify the cancer susceptibility. For example, Wirtenberger et al. found that polymorphisms rs4645959 A > G within the N-terminal domain of MYC led to an amino acid transition from Asn to Ser; and the heterozygous carriers of the Asn11Ser had increased breast cancer risk (46). SNPs in cis regulators of transcription of MYC gene may change its germline expression levels and contribute to cancer susceptibility. The rs13281615 located in the non-coding region near the MYC has been reported to associate with the risk of colorectal cancer (47), prostate cancer (48), breast cancer (49). The rs6983267 affects the binding of MYC with transcription factor 7-like 2 (TCF7L2) and TCF4 (50), and it is proved to up-regulate the transcription of MYC by Takatsuno et al. (51). And the rs6983267 has been reported to modify the cancer susceptibility, such as gastric cancer (52). Guo et al. demonstrated that rs4645948 located in 5' UTR of the MYC gene increased the transcriptional activity of MYC, and thus increased risk of developing nasopharyngeal carcinoma (53). These studies mention above all indicated that SNPs in the MYC gene may affect its expression and function, then contribute to the cancer susceptibility.
The rs4645943 C > T and rs2070583 A > G polymorphisms are located in 5’ UTR and 3' UTR region of the MYC gene, respectively, two crucial regions for regulating its expression. The polymorphism rs4645943 C > T has been reported to be associated with prostate cancer risk (28). However, no study has reported the association between this polymorphism and the WT risk. Moreover, there is no association study regarding rs2070583 A > G and cancer susceptibility. In this current case-control study, we attempt to investigate the association between the selected SNPs in the regulatory region of MYC gene and WT susceptibility in a Chinese population. However, no association between two selected polymorphisms and WT risk was found. The two selected polymorphisms might not affect the MYC gene expression and function, and therefore would not modify the susceptibility to WT.
Several limitations should be mentioned in this study. First, the sample size is moderate. A larger sample size would increase the statistical power and the credibility of the conclusions. Second, only two polymorphisms of the MYC gene were evaluated, other potentially functional MYC SNPs should be investigated. Third, selection bias is inevitable, resulting from this hospital-based study design. Therefore, the study population may not fully represent the general population.
In summary, our results showed that both two MYC gene associated polymorphisms (rs4645943 C > T, and rs2070583 A > G) were not associated with WT risk in Chinese population. Future studies with larger sample size comprising different ethnicities should be performed to confirm our conclusion.
Acknowledgments
Funding: This study was funded by grants from the National Natural Science Foundation of China (No: 81803320), the Pearl River S&T Nova Program of Guangzhou (No: 201710010086), the Science and Technology Project of Guangzhou (No: 201804010037) and the Science and Technology Planning Project of Guangdong Province (No: 2016A020215009).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: Our study design received approval from the each institutional review board of all participating hospitals. Written informed consent was obtained from all patients. The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Ko EY, Ritchey ML. Current management of Wilms' tumor in children. J Pediatr Urol 2009;5:56-65. [Crossref] [PubMed]
- Chu A, Heck JE, Ribeiro KB, et al. Wilms' tumour: a systematic review of risk factors and meta-analysis. Paediatr Perinat Epidemiol 2010;24:449-69. [Crossref] [PubMed]
- Breslow NE, Olson J, Moksness J, et al. Familial Wilms' tumor: a descriptive study. Med Pediatr Oncol 1996;27:398-403. [Crossref] [PubMed]
- Bao PP, Li K, Wu CX, et al. Recent incidences and trends of childhood malignant solid tumors in Shanghai, 2002-2010. Zhonghua Er Ke Za Zhi 2013;51:288-94. [PubMed]
- Rivera MN, Haber DA. Wilms' tumour: connecting tumorigenesis and organ development in the kidney. Nat Rev Cancer 2005;5:699-712. [Crossref] [PubMed]
- Charlton J, Williams RD, Sebire NJ, et al. Comparative methylome analysis identifies new tumour subtypes and biomarkers for transformation of nephrogenic rests into Wilms tumour. Genome Med 2015;7:11. [Crossref] [PubMed]
- Dome JS, Graf N, Geller JI, et al. Advances in Wilms Tumor Treatment and Biology: Progress Through International Collaboration. J Clin Oncol 2015;33:2999-3007. [Crossref] [PubMed]
- Szychot E, Apps J, Pritchard-Jones K. Wilms' tumor: biology, diagnosis and treatment. Transl Pediatr 2014;3:12-24. [PubMed]
- Malogolowkin M, Cotton CA, Green DM, et al. Treatment of Wilms tumor relapsing after initial treatment with vincristine, actinomycin D, and doxorubicin. A report from the National Wilms Tumor Study Group. Pediatr Blood Cancer 2008;50:236-41. [Crossref] [PubMed]
- Huff V. Wilms tumor genetics. Am J Med Genet 1998;79:260-7. [Crossref] [PubMed]
- Pritchard-Jones K, Moroz V, Vujanic G, et al. Treatment and outcome of Wilms' tumour patients: an analysis of all cases registered in the UKW3 trial. Ann Oncol 2012;23:2457-63. [Crossref] [PubMed]
- Deng C, Dai R, Li X, et al. Genetic variation frequencies in Wilms' tumor: A meta-analysis and systematic review. Cancer Sci 2016;107:690-9. [Crossref] [PubMed]
- Maiti S, Alam R, Amos CI, et al. Frequent association of beta-catenin and WT1 mutations in Wilms tumors. Cancer Res 2000;60:6288-92. [PubMed]
- Rivera MN, Kim WJ, Wells J, et al. An X chromosome gene, WTX, is commonly inactivated in Wilms tumor. Science 2007;315:642-5. [Crossref] [PubMed]
- Ruteshouser EC, Robinson SM, Huff V. Wilms tumor genetics: mutations in WT1, WTX, and CTNNB1 account for only about one-third of tumors. Genes Chromosomes Cancer 2008;47:461-70. [Crossref] [PubMed]
- Rapley EA, Barfoot R, Bonaiti-Pellie C, et al. Evidence for susceptibility genes to familial Wilms tumour in addition to WT1, FWT1 and FWT2. Br J Cancer 2000;83:177-83. [Crossref] [PubMed]
- Yost S, de Wolf B, Hanks S, et al. Biallelic TRIP13 mutations predispose to Wilms tumor and chromosome missegregation. Nat Genet 2017;49:1148-51. [Crossref] [PubMed]
- Wegert J, Ishaque N, Vardapour R, et al. Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high-risk blastemal type Wilms tumors. Cancer Cell 2015;27:298-311. [Crossref] [PubMed]
- Walz AL, Ooms A, Gadd S, et al. Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer Cell 2015;27:286-97. [Crossref] [PubMed]
- Ciceri S, Gamba B, Corbetta P, et al. Genetic and epigenetic analyses guided by high resolution whole-genome SNP array reveals a possible role of CHEK2 in Wilms tumour susceptibility. Oncotarget 2018;9:34079-89. [Crossref] [PubMed]
- Fu W, Zhu J, Xiong SW, et al. BARD1 Gene Polymorphisms Confer Nephroblastoma Susceptibility. EBioMedicine 2017;16:101-5. [Crossref] [PubMed]
- Fu W, Liu GC, Zhao Z, et al. The correlation between LIN28B gene potentially functional variants and Wilms tumor susceptibility in Chinese children. J Clin Lab Anal 2018.32. [PubMed]
- Fu W, Li L, Xiong SW, et al. miR-423 rs6505162 C>A polymorphism contributes to decreased Wilms tumor risk. J Cancer 2018;9:2460-5. [Crossref] [PubMed]
- Zhu J, Fu W, Jia W, et al. Association between NER Pathway Gene Polymorphisms and Wilms Tumor Risk. Mol Ther Nucleic Acids 2018;12:854-60. [Crossref] [PubMed]
- Miller DM, Thomas SD, Islam A, et al. c-Myc and cancer metabolism. Clin Cancer Res 2012;18:5546-53. [Crossref] [PubMed]
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer 2008;8:976-90. [Crossref] [PubMed]
- Fernandez PC, Frank SR, Wang L, et al. Genomic targets of the human c-Myc protein. Genes Dev 2003;17:1115-29. [Crossref] [PubMed]
- Haiman CA, Patterson N, Freedman ML, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet 2007;39:638-44. [Crossref] [PubMed]
- Terunuma A, Putluri N, Mishra P, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest 2014;124:398-412. [Crossref] [PubMed]
- De Baets G, Van Durme J, Reumers J, et al. SNPeffect 4.0: on-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res 2012;40:D935-9. [Crossref] [PubMed]
- Noreen M, Arshad M. Association of TLR1, TLR2, TLR4, TLR6, and TIRAP polymorphisms with disease susceptibility. Immunol Res 2015;62:234-52. [Crossref] [PubMed]
- Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet 2007;39:989-94. [Crossref] [PubMed]
- Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet 2007;39:631-7. [Crossref] [PubMed]
- Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007;447:1087-93. [Crossref] [PubMed]
- Fu W, Zhuo Z, Hua RX, et al. Association of KRAS and NRAS gene polymorphisms with Wilms tumor risk: a four-center case-control study. Aging (Albany NY) 2019;11:1551-63. [Crossref] [PubMed]
- Liu P, Zhuo Z, Li W, et al. TP53 rs1042522 C>G polymorphism and Wilms tumor susceptibility in Chinese children: a four-center case-control study. Biosci Rep 2019;39:BSR20181891. [Crossref] [PubMed]
- Zhou H, Zhuo Z, Chen S, et al. Polymorphisms in MYCN gene and neuroblastoma risk in Chinese children: a 3-center case-control study. Cancer Manag Res 2018;10:1807-16. [Crossref] [PubMed]
- Gearhart J, Pashos EE, Prasad MK. Pluripotency redux--advances in stem-cell research. N Engl J Med 2007;357:1469-72. [Crossref] [PubMed]
- Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 2005;6:635-45. [Crossref] [PubMed]
- van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 2010;10:301-9. [Crossref] [PubMed]
- Kim J, Woo AJ, Chu J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 2010;143:313-24. [Crossref] [PubMed]
- Dejure FR, Eilers M. MYC and tumor metabolism: chicken and egg. EMBO J 2017;36:3409-20. [Crossref] [PubMed]
- Casey SC, Baylot V, Felsher DW. MYC Master Regulator of Immune Privilege. Trends Immunol 2017;38:298-305. [PubMed]
- Batistatou A, Kotoula V, Bobos M, et al. Correlation of MYC Gene and Protein Status With Breast Cancer Subtypes and Outcome of Patients Treated With Anthracycline-Based Adjuvant Chemotherapy. Pooled Analysis of 2 Hellenic Cooperative Group Phase III Trials. Clin Breast Cancer 2018;18:53-62.e3. [Crossref] [PubMed]
- Kim EY, Kim A, Kim SK, et al. MYC expression correlates with PD-L1 expression in non-small cell lung cancer. Lung Cancer 2017;110:63-7. [Crossref] [PubMed]
- Wirtenberger M, Hemminki K, Forsti A, et al. c-MYC Asn11Ser is associated with increased risk for familial breast cancer. Int J Cancer 2005;117:638-42. [Crossref] [PubMed]
- Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet 2007;39:984-8. [Crossref] [PubMed]
- Meyer A, Schurmann P, Ghahremani M, et al. Association of chromosomal locus 8q24 and risk of prostate cancer: a hospital-based study of German patients treated with brachytherapy. Urol Oncol 2009;27:373-6. [Crossref] [PubMed]
- Pei YL, Zhang HL, Han HG. Polymorphism of 8q24 rsl3281615 and breast cancer risk: a meta-analysis. Tumour Biol 2013;34:421-8. [Crossref] [PubMed]
- Pomerantz MM, Ahmadiyeh N, Jia L, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet 2009;41:882-4. [Crossref] [PubMed]
- Takatsuno Y, Mimori K, Yamamoto K, et al. The rs6983267 SNP is associated with MYC transcription efficiency, which promotes progression and worsens prognosis of colorectal cancer. Ann Surg Oncol 2013;20:1395-402. [Crossref] [PubMed]
- Guo Y, Fang J, Liu Y, et al. Association between polymorphism rs6983267 and gastric cancer risk in Chinese population. World J Gastroenterol 2011;17:1759-65. [Crossref] [PubMed]
- Guo Z, Wang Y, Zhao Y, et al. A Functional 5'-UTR Polymorphism of MYC Contributes to Nasopharyngeal Carcinoma Susceptibility and Chemoradiotherapy Induced Toxicities. J Cancer 2019;10:147-55. [Crossref] [PubMed]


