
Deep learning-based automated diagnosis of fungal keratitis with in vivo confocal microscopy images
Introduction
Fungal keratitis (FK) is a serious corneal infection caused by fungal organisms and is the main cause of vision loss and blindness, especially in developing countries (1,2). Early diagnosis and treatment are the key to preventing severe complications, such as corneal perforation, anterior chamber empyema, endophthalmitis and blindness (3-5). At present, the diagnosis of FK mainly relies on the following methods: slit-lamp examination, corneal scraping microscopy examination, fungal culture, tissue biopsy, confocal microscopy and polymerase chain reaction (PCR) (6,7). These methods have their own advantages in diagnosing FK from different aspects, along with some shortcomings or limits. For example, doctors can observe only the superficial signs by slit-lamp examination to make an initial diagnosis (8,9), corneal scraping microscopy examination and tissue biopsy are invasive tests that may cause secondary infection (10), fungal culture takes a relatively long time and may produce negative results, which is difficult for early diagnosis (3), and the high cost of PCR limits its extensive clinical application (7,11,12).
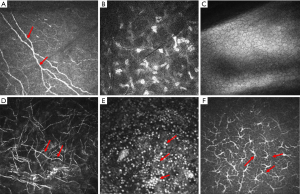
In vivo confocal microscopy (IVCM) is a noninvasive imaging modality that can present the direct visualization of potential causative pathogens in real time (13,14). It uses laser as a light source to scan organisms point by point and line by line to obtain high-resolution optical cross-sectional images of thick specimens (15-17). IVCM also overcomes the shortcomings of traditional optical microscopes by using spatial conjugated pinholes to block out-of-focal plane rays to obtain clear images (18,19). The images collected via IVCM show cell morphology, so it can be used to diagnose FK at the cellular level (20,21). As shown in Figure 1, the confocal microscopy images in the normal cornea show a clear background with corneal nerve fibers and other structures, however, in the pathological cornea, various hyphae and inflammatory cells. Moreover, especially in diabetic patients, the decrease in the number and continuity of nerve fibers can also seriously interfere with the accuracy of diagnosis, because the morphology of the hyphae are the most important basis for clinical diagnosis (22). Therefore, identifying key structures in different images is an important and challenging task for this subject. Furthermore, confocal microscopy is an imaging diagnostic technique that completely relies on the clinical experience and subjective judgment of the attending doctors (23). Moreover, doctors have to screen, observe and analyze plenty of images acquired by the confocal microscope continuous tomography scan, which is time-consuming and laborious work (14).
With the rapid development of computer and image processing technologies, the deep learning method is a revolutionary advancement in image recognition that has been used to analyze various diseases, such as diabetic retinopathy age-related macular degeneration, glaucoma and optic neuropathies (24-26). Deep learning methods are family of machine-learning techniques, that contains multiple hidden-layer networks that allow the production of highly accurate algorithms in diagnosing diseases from medical images with accuracy comparable to that of human experts (24,27-29). Moreover, Deep Residual Learning for Image Recognition (ResNet) is a powerful deep learning method which have been applied in diagnosis widely, such as for the detection of glaucoma in both highly myopic and non-highly myopic eyes (24). Machine learning has been reportedly applied to automatically diagnose FK. For example, an automatic diagnostic algorithm based on adaptive random backpropagation (ARBP) and support vector machine (SVM) achieves 99.74% accuracy from IVCM images. However, there are no reports on the diagnosis of FK with the use of a deep learning method or ResNet so far. Therefore, this study aims to construct a model based on ResNet automatically to screen FK from IVCM images automatically and to use an independent dataset to validate its diagnostic performance.
Methods
This study followed the tenets of the Helsinki Declaration. Because of the retrospective nature and fully anonymized use of images in this study, The Review Board of the People’s Hospital of Guangxi Zhuang Autonomous Region indicated that informed consent was not required.
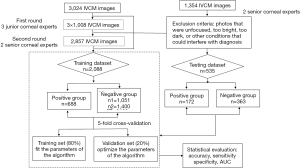
The training dataset was prepared using photographs recorded with IVCM (HRT III/RCM Heidelberg Engineering, Germany) between November 2018 and February 2019 at Guangxi Zhuang Autonomous Region People’s Hospital, China. All images were recorded as JPEG format with a resolution of 384×384 pixels. These keratitis patients were confirmed to have fungal infections by fungal culture and were cured by certain antibiotics or antifungal drugs. After excluding images that were unfocused, too bright, too dark, or had other conditions that could interfere with diagnosis, a total of 2,088 IVCM images were included in the training dataset. They were screened, observed and analyzed by five experts specializing in corneal disease. All experts were required to make their own diagnosis independently with no communication.
The image preparation process was as follows. First, IVCM images were randomly assigned to three junior corneal experts for initial screening and labeling. Each expert had to review 1,008 images. After excluding images according to the criteria, 2,857 images were included in the next round of screening. Second, the other two corneal experts were invited to confirm the labeling results. If the diagnoses of the first and second round experts were inconsistent, the image was submitted to the highest level of corneal expertise for a final decision. Ultimately, 2,088 IVCM images were included in the training dataset. A total of 688 images of species with hyphae were classified into the positive group, while 1,051 images were classified into the negative group which included 426 images of normal corneal tissue and 625 images of corneal tissue with inflammatory cells and/or activated dendritic cells. Moreover, 349 corneal confocal images from diabetic patients were added to the negative group.
The testing dataset was filtered between March 2019 and June 2019 at Guangxi Zhuang Autonomous Region People’s Hospital, China. The basic requirements of all images in the testing dataset were consistent with those of the training dataset. The positive group contained 172 images, and the negative group contained 363 images (normal corneal tissue, from normal individuals and patients with diabetes as well as corneal tissue with inflammatory cells and/or activated dendritic cells). The diagnosis of FK was performed independently by two experts with extensive clinical experience. If the diagnoses of the two experts were inconsistent, the images were confirmed by the corneal expert. Figure 2 shows the detailed model building process.
Development of the intelligent system
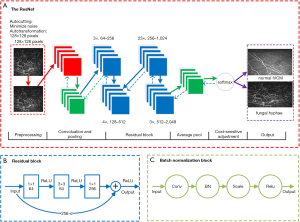
In this study, we used 101 layer ResNet, which is an enhanced deep learning algorithm based on a convolutional neural network (CNN) (30). ResNet could gain higher accuracy from the deeper network than CNN when classing images. When testing the model, the output of our model was whether the image was FK or not. The architecture of the ResNet used in this paper is shown in Figure 3. It consists of 33 residual blocks. Each block is composed of three convolutional layers (1×1, 3×3 and 1×1 convolutions), which with different numbers of channels. The 1×1 convolutions are responsible for reducing and increasing the dimensions of the channels, and the 3×3 convolution is the main processing unit. Overall, the ResNet framework contains 99 convolutional layers and 2 pooling layers. Each pixel on each output channel is computed using the convolution between the three-dimensional kernel and the corresponding pixels across the three input channels (31).
Statistical evaluation
During the training process, the training dataset was randomly divided into five subsets, the deep learning method was trained using four of the five arms, and the remaining arm was used for calculating the diagnostic accuracy. This process was iterated 5 times so that each of the five arms was used as a validation dataset once.
The testing dataset was used to perform external validation. And the receiver operating characteristic (ROC) curve and the area under the ROC curve (AUC) with 95% confidence intervals (CIs) were adopted to assess the performance of the algorithm (25). The ROC curve plots the true positive rate (TPR) against the false positive rate (FPR). The best possible classification method would yield the point (0.1) of the ROC space, which represents 100% sensitivity and 100% specificity (32).
Results
This study built a novel automated-feature learning intelligent system based on a deep learning algorithm to detect FK in IVCM images. The testing dataset included 172 images of fungal hyphae and 363 IVCM images in the negative group (239 images of normal corneal tissue and 124 images of corneal tissue with inflammatory cells and/or activated dendritic cells).
When there were 1,051 images in the negative group in the training dataset, 515 images were correctly diagnosed in the testing dataset, of which 58 images were diagnosed as fungal images and 357 images were diagnosed as nonfungal images. Twenty images were misdiagnosed, of which 6 images were misdiagnosed as fungal images and 14 images were misdiagnosed as nonfungal images (panel A of Figure 4). In the testing dataset, the system showed an accuracy of 0.9626 with a specificity of 0.9834 and a sensitivity of 0.9186 (panel A of Figure 5). The performance of this system was evaluated by a ROC curve (panel A of Figure 6). The AUC value with ResNet was 0.9875 (95% CI, 0.976–0.991).
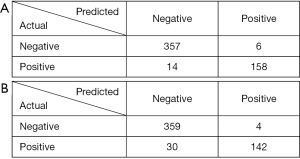
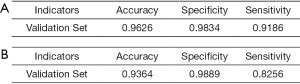
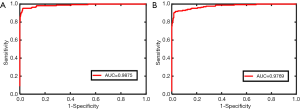
When 349 images of diabetic patients were added to the training dataset, the testing dataset showed different results with 501 images diagnosed correctly and 34 misdiagnosed (panel B of Figure 4). The 501 images diagnosed correctly included 142 images with fungus and 359 without. The 34 misdiagnosed images consisted of 4 with fungus and 30 without. An accuracy of 0.9364 with a specificity of 0.9889 and a sensitivity of 0.8256 was obtained in the testing dataset (panel B of Figure 5). The ROC curve obtained for the testing dataset is shown in panel B of Figure 6. The AUC value with ResNet was 0.9769 (95% CI, 0.976–0.991).
Discussion
Diagnosing FK quickly is a very important and challenging task because microbial culture often takes a relatively long time and may produce negative results. IVCM, a noninvasive imaging modality, has been widely used to diagnose FK at the cellular level. However, the most annoying problem is that assessing a large number IVCM images is time consuming. In this study, we constructed a novel automated-feature learning intelligent system to detect the FK with IVCM images. The system could not only help doctors diagnose FK quickly but also monitor the presence of fungal residues during the treatment process. The deep learning method has been indicated to have scientific merit by many reports (30,33,34), and this system based on ResNet also exhibited excellent performance (accuracy, 0.9626; sensitivity, 0.9186; and specificity, 0.9834) for the diagnosis of FK. Even in diabetic population, our system based on ResNet showed high diagnostic power (accuracy, 0.9364; sensitivity, 0.8256; and specificity, 0.9889) in diagnosing FK, which was validated in an independent testing dataset.
The ResNet deep network in our study used 101 conventional layers, and could extract more complex and detailed features from various IVCM images. It also showed a more powerful performance than the traditional model based on CNN because the influence resulting from gradient decline and divergence had been decreased. It is more difficult to diagnose FK correctly from IVCM images of diabetic patients because of the decrease in the number and continuity of nerve fibers in the corneal tissue of diabetic patients. The accuracy of the intelligent system based on a deep learning algorithm for diagnosing FK with IVCM images would also be influenced. Our system also proved this finding, as the testing dataset’s accuracy decreased from 0.9626 to 0.9364 when 349 images of diabetic patients were added to the training dataset. This result implied that the algorithm still needed to be improved in the diagnosis of FK in diabetic patients. Though the accuracy of this system decreased when images from diabetic patients were included, the model including images of diabetic patients is more realistic because diabetic patients are common in the clinic. Furthermore, all images in our study, regardless of the training dataset or testing dataset, are actual photographs without artificial feature extraction, which would avoid misalignment and misclassification (35,36).
Most importantly, this is the first attempt to build a model with external verification to diagnose FK with IVCM images, and all images of FK in our study were proven by fungal culture. The accuracy of 0.9626 in our study is not as high as that previously reported (accuracy of 0.9974) (9). However, unlike other studies, the accuracy of this system is the result of an external verification set which is of more clinical significance.
In this study, we developed a robust diagnostic system for FK detection within a largescale data set. The results indicated the powerful performance of our computer-aided model in providing efficient, low-cost, and objective FK diagnostics, which relieves clinicians from examining and grading images manually one by one. However, in the medical diagnostic field, an accuracy of 0.9626 is not sufficient for precise treatment. It can be considered only an auxiliary diagnostic method to provide a reference for timely treatment. Improving the clarity of the images, increasing the number and variety of images, and optimizing the parameters of the algorithm can improve the accuracy of the model; however, microbial culture is still the diagnostic gold standard. Further research is warranted to check the performance of the model in different environments with different devices, such as slit-lamp microscopy and camera systems. Moreover, this system could judge only whether there is fungal infection or not and could not identify fungal subtypes and the severity of infection. In addition, the visualization of the exact features seen and identified by the system still needs to be realized, which could help doctors, patients and health care providers understand its learning process better (37). In future work, we aim to perform multicenter research and establish a larger database of FK to improve the accuracy and broaden the applicability of the system and to explore suitable deep learning algorithms to achieve visualization.
In conclusion, a deep learning method was developed to automatically screen FK from IVCM images that could reduce the workload of clinicians. With the increase in samples and optimization of the algorithms, artificial intelligence systems will have a significant clinical impact and may completely change the current mode of disease diagnosis.
Acknowledgments
Funding: This study was supported by the National Key Research and Development Program (2018YFC0116500); Guangxi Promotion of Appropriate Health Technologies Project (No. S2019084).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Haotian Lin and Limin Yu) for the series “Medical Artificial Intelligent Research” published in Annals of Translational Medicine. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.03.134). The series “Medical Artificial Intelligent Research” was commissioned by the editorial office without any funding or sponsorship. All authors report grants from The National Key Research and Development Program (2018YFC0116500), grants from Guangxi Promotion of Appropriate Health Technologies Project (No. S2019084), during the conduct of the study.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of the People’s Hospital of Guangxi Zhuang Autonomous Region (KY-SY-2020-1). Because of fully anonymized usage of images in this study, the review board indicated that the informed consent was not required.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li X, Yuan M, Yin R, et al. Histone deacetylase inhibitor attenuates experimental fungal keratitis in mice. Sci Rep 2019;9:9859. [Crossref] [PubMed]
- Alshehri JM, Caballero-Lima D, Hillarby MC, et al. Evaluation of Corneal Cross-Linking for Treatment of Fungal Keratitis: Using Confocal Laser Scanning Microscopy on an Ex Vivo Human Corneal Model. Invest Ophthalmol Vis Sci 2016;57:6367-73. [Crossref] [PubMed]
- Thomas PA. Fungal infections of the cornea. Eye 2003;17:852-62. [Crossref] [PubMed]
- Kheirkhah A, Syed ZA, Satitpitakul V, et al. Sensitivity and Specificity of Laser-Scanning In Vivo Confocal Microscopy for Filamentous Fungal Keratitis: Role of Observer Experience. Am J Ophthalmol 2017;179:81-9. [Crossref] [PubMed]
- Chew SJ, Beuerman RW, Assouline M, et al. Early diagnosis of infectious keratitis with in vivo real time confocal microscopy. CLAO J 1992;18:197-201. [PubMed]
- Belliappa S, Hade J, Kim S, et al. Surgical Outcomes in Cases of Contact Lens-Related Fusarium Keratitis. Eye Contact Lens 2010;36:190-4. [Crossref] [PubMed]
- Goh JWY, Harrison R, Hau S, et al. Comparison of In Vivo Confocal Microscopy, PCR and Culture of Corneal Scrapes in the Diagnosis of Acanthamoeba Keratitis. Cornea 2018;37:480-5. [Crossref] [PubMed]
- Mahmoudi S, Masoomi A, Ahmadikia K, et al. Fungal keratitis: An overview of clinical and laboratory aspects. Mycoses 2018;61:916-30. [Crossref] [PubMed]
- Wu X, Qiu Q, Liu Z, et al. Hyphae Detection in Fungal Keratitis Images With Adaptive Robust Binary Pattern. IEEE Access 2018;6:13449-60.
- Du R, Wu J, Ma JX, et al. Establishment of rabbit fungal (aspergillus) keratitis model and corneal examination with confocal microscope. J Fourth Mil Med Univ 2006;27:1925-28.
- Thomas PA, Teresa PA, Theodore J, et al. PCR for the molecular diagnosis of mycotic keratitis. Expert Rev Mol Diagn 2012;12:703-18. [Crossref] [PubMed]
- Ferrer C, Alio JL. Evaluation of molecular diagnosis in fungal keratitis. Ten years of experience. J Ophthalmic Inflamm Infect 2011;1:15-22. [Crossref] [PubMed]
- Labbé A, Khammari C, Dupas B, et al. Contribution of In Vivo Confocal Microscopy to the Diagnosis and Management of Infectious Keratitis. Ocul Surf 2009;7:41-52. [Crossref] [PubMed]
- Niederer RL, McGhee CN. Clinical in vivo confocal microscopy of the human cornea in health and disease. Prog Retin Eye Res 2010;29:30-58. [Crossref] [PubMed]
- Patel DV, Zhang J, McGhee CN. In vivo confocal microscopy of the inflamed anterior segment a review of clinical and research applications. Clin Exp Ophthalmol 2019;47:334-45. [Crossref] [PubMed]
- Matsumoto Y, Dogru M, Sato EA, et al. The application of in vivo confocal scanning laser microscopy in the management of Acanthamoeba keratitis. Mol Vis 2007;13:1319-26. [PubMed]
- Vaddavalli PK, Garg P, Sharma S, et al. Confocal microscopy for Nocardia keratitis. Ophthalmology 2006;113:1645-50. [Crossref] [PubMed]
- Chidambaram JD, Prajna NV, Larke NL, et al. Prospective Study of the Diagnostic Accuracy of the In Vivo Laser Scanning Confocal Microscope for Severe Microbial Keratitis. Ophthalmology 2016;123:2285-93. [Crossref] [PubMed]
- Hamrah P, Sahin A, Dastjerdi MH, et al. Cellular changes of the corneal epithelium and stroma in herpes simplex keratitis: an in vivo confocal microscopy study. Ophthalmology 2012;119:1791-7. [Crossref] [PubMed]
- Chidambaram JD, Prajna NV, Palepu S, et al. In Vivo Confocal Microscopy Cellular Features of Host and Organism in Bacterial, Fungal, and Acanthamoeba Keratitis. Am J Ophthalmol 2018;190:24-33. [Crossref] [PubMed]
- Cavanagh HD, Jester JV, Essepian J, et al. Confocal microscopy of the living eye. CLAO J 1990;16:65-73. [PubMed]
- O'Donnell C, Efron N. Corneal Endothelial Cell Morphometry and Corneal Thickness in Diabetic Contact Lens Wearers. Optom Vis Sci 2004;81:858-62. [Crossref] [PubMed]
- Hau SC, Dart JK, Vesaluoma M, et al. Diagnostic accuracy of microbial keratitis with in vivo scanning laser confocal microscopy. Br J Ophthalmol 2010;94:982-7. [Crossref] [PubMed]
- Shibata N, Tanito M, Mitsuhashi K, et al. Development of a deep residual learning algorithm to screen for glaucoma from fundus photography. Sci Rep 2018;8:14665. [Crossref] [PubMed]
- Zhang K, Liu X, Liu F, et al. An Interpretable and Expandable Deep Learning Diagnostic System for Multiple Ocular Diseases: Qualitative Study. J Med Internet Res 2018;20:e11144. [Crossref] [PubMed]
- Hwang DK, Hsu CC, Chang KJ, et al. Artificial intelligence-based decision-making for age-related macular degeneration. Theranostics 2019;9:232-45. [Crossref] [PubMed]
- Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. [Crossref] [PubMed]
- O'Bryhim BE, Apte RS, Kung N, et al. Association of Preclinical Alzheimer Disease With Optical Coherence Tomographic Angiography Findings. JAMA Ophthalmol 2018;136:1242-8. [Crossref] [PubMed]
- Lu W, Tong Y, Yu Y, et al. Deep Learning-Based Automated Classification of Multi-Categorical Abnormalities From Optical Coherence Tomography Images. Transl Vis Sci Technol 2018;7:41. [Crossref] [PubMed]
- Qiu Q, Liu Z, Zhao Y, et al. Automatic Detecting Cornea Fungi Based on Texture Analysis. 2016 IEEE International Conference on Smart Cloud (SmartCloud)2016. p. 214-7.
- Wu X, Huang Y, Liu Z, et al. Universal artificial intelligence platform for collaborative management of cataracts. Br J Ophthalmol 2019;103:1553-60. [Crossref] [PubMed]
- Oh JH, Craft J, Al Lozi R, et al. A Bayesian network approach for modeling local failure in lung cancer. Phys Med Biol 2011;56:1635-51. [Crossref] [PubMed]
- Wu X, Tao Y, Qiu Q, et al. Application of image recognition-based automatic hyphae detection in fungal keratitis. Australas Phys Eng Sci Med 2018;41:95-103. [Crossref] [PubMed]
- Sharif MS, Qahwaji R, Ipson S, et al. Medical image classification based on artificial intelligence approaches: A practical study on normal and abnormal confocal corneal images. Applied Soft Computing 2015;36:269-82. [Crossref]
- Li Z, He Y, Keel S, et al. Efficacy of a Deep Learning System for Detecting Glaucomatous Optic Neuropathy Based on Color Fundus Photographs. Ophthalmology 2018;125:1199-206. [Crossref] [PubMed]
- Yang M, Zhang L, Shiu SC, et al. Robust kernel representation with statistical local features for face recognition. IEEE Trans Neural Netw Learn Syst 2013;24:900-12. [Crossref] [PubMed]
- Wong TY, Bressler NM. Artificial Intelligence With Deep Learning Technology Looks Into Diabetic Retinopathy Screening. JAMA 2016;316:2366-7. [Crossref] [PubMed]


