
Real-time estimation of the reproduction number of the novel coronavirus disease (COVID-19) in China in 2020 based on incidence data
Introduction
The novel coronavirus disease (COVID-19) cases have appeared in Wuhan, China since December 2019, while the causes of the first 4 cases are related to the South China Seafood Wholesale Market in Wuhan. Through the large-scale travelling activities, the COVID-19 has soon spread to other Chinese cities and other countries (Japan, Singapore, Thailand, etc.) (1,2). The COVID-19 could result in main symptoms including malaise, fever, dry cough, nasal congestion, diarrhea, and dyspnea, etc. (3). By examining the genome sequence of the virus, the pathogen of COVID-19 was found phylogenetically similar to the severe acute respiratory syndrome coronavirus (SARS-CoV), which caused outbreaks in China in 2003. Thus, the pathogen of COVID-19 was named as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the World Health Organization (WHO).
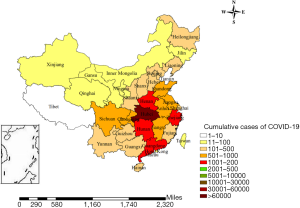
In order to reduce the risk of infection, the local government in Wuhan issued a notice that all within-city and cross-border public transportations were suspended as of January 23 (effected on 10 AM, GMT+8). Since then, the other 12 cities in Hubei province also implemented similar public traffic control measures. On January 30, the WHO declared that the COVID-19 outbreak constitutes a Public Health Emergency of International Concern (PHEIC) (4). Recently, studies have shown that the disease could be transmitted from person to person through droplets and contact (5). As of February 16, 2020, China has accumulated 70,548 confirmed cases, 1,770 dead cases, and 10,844 cured and discharged cases, of which Hubei province has the most severe epidemic situation, with a total of 58,182 confirmed cases (41,152 in Wuhan), accounting for 82.47% of the total number of confirmed cases nationwide, and a total of 1,696 death cases (1,309 cases in Wuhan), accounting for 98.82% of the national death cases. The provinces with more than 1,000 confirmed cases in China are Hubei, Guangdong, Zhejiang, Henan, and Hunan (Figure 1).
The quantification of transmissibility-during epidemic is essential to design and adjust public health responses. Transmissibility can be measured by the reproduction number, the average number of secondary cases caused by an infected individual (6-9). Knowledge of the reproductive number is crucial for understanding the dynamics of any infectious disease, and these should be reevaluated as the pandemic progresses in space and time. Recently, the estimations of reproduction number of COVID-19 have been discussed in (1,2,10-14). The existence of these different findings is due to analyzing the problem from different perspectives. These authors selected datasets at different intervals, and the methods and parameters are also different. These different modeling methods have a bit varied estimates of reproduction numbers and also reflect the spread of coronavirus outbreaks and the dynamics of case reports differently. These findings provide important parameters for further analysis, including assessing the impact of control measures and predicting the spread of future infections. Wu et al. (2) carried out a modeling study and estimated that the basic reproductive number for COVID-19 at 2.68 (95% CI: 2.47–2.86) from 1 December 2019 to 25 January 2020. Based on the cumulative number of reported cases data from 10 January 2020 to 22 January 2020 of Wuhan and Hubei province, Tang et al. (10) have estimated the basic reproduction numbers of Wuhan using likelihood-based and model-based approaches. The mean control reproduction number was estimated to be as high as 6.47 (95% CI: 5.71–7.23). Zhao et al. (11) adopted a data-driven modelling analysis of the Early Outbreak to estimate the number of unreported cases and the basic reproduction number of COVID-19 in Wuhan. They modeled the epidemic curve of COVID-19 cases, in mainland China from 1 December 2019 to 24 January 2020 through the exponential growth. After taking into account of the effect of under-reporting, the R0 was estimated at 2.56 (95% CI: 2.49−2.63). Meanwhile, Zhao et al. (12) modeled the epidemic curve of COVID-19 cases time series, in mainland China from January 10 to January 24, 2020, through the exponential growth. They estimated R0 by using the serial intervals (SI) of two other well-known coronavirus diseases, MERS and SARS, as approximations for the truly unknown SI. Their results showed that the mean R0 ranges from 2.24 (95% CI: 1.96–2.55) to 3.58 (95% CI: 2.89–4.39) with 2-fold to 8-fold increase in the reporting rate. Li et al. (1) collected information on demographic characteristics, exposure history, and illness timelines of laboratory-confirmed cases of NCIP (with 425 confirmed patients), that had been reported from January 1, 2020, to January 22, 2020. Using the data, they analyzed the early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. In the early period of exponential growth, they estimated the basic reproductive number was 2.2 (95% CI: 1.4 to 3.9). The dynamics transmission model by Shen et al. (13) computed a basic reproduction number of 4.71 (95% CI: 4.50–4.92), which decreased to 2.08 (95% CI: 1.99–2.18) on January 22nd, 2020. Finally, Riou et al. (14) computed a reproduction number of 2.2 (90% CI: 1.4–3.8) by using a stochastic model.
With the implementation of COVID-19 control measures, the disease transmissibility was likely to change as well. Based on the surveillance data of COVID-19, we calculated the time-varying reproduction number, evaluated its changing dynamics from January 16 to February 16. Meanwhile, the effects of the COVID-19 control measures were evaluated by comparing the reproduction numbers in different periods of time.
Methods
We collected the data of the confirmed COVID-19 cases from the National Health Commission of the People’s Republic of China from January 10 to February 16, 2020 (15). All cases were positively diagnosed based on the laboratory nucleic acid test, which is not excluded from the clinical diagnosis case. The time series plot of the daily number of COVID-19 cases is shown in Figure 2.

In this study, we divided the timeline of the outbreak into three stages:
- Stage I was from January 16 to January 22 before the ‘city lockdown’ was implemented in Wuhan;
- Stage II was from January 23 to January 30 before the WHO announced PHEIC; and
- Stage III was the time period after the PHEIC announced by WHO from January 31st to February 16.
We estimated the time-varying reproduction number, also known as the instantaneous reproduction number, by using the renewable equation proposed in Wallinga and Teunis (16). The relative likelihood
|
| [1] |
The effective reproduction number for case j is the sum of all cases i, weighted by the relative likelihood that case i has been infected by case j:
|
| [2] |
Additional detail on the derivation of these equations is provided in (17-19). The analysis was carried out by using R (version 3.6.0).
The SI is defined as the time between infections in a consecutive chain of transmission (20). The SI determines, along with the reproduction number, the growth rate of an epidemic curve. In You et al.’s study (21), the SI of COVID-19 was estimate at 4.4 days with a SD at 3.17 days based on 71 records. Thus, in this study, the SI is governed by a Gamma distribution with a mean of 4.4 days and a standard deviation of 3.17 days. Additionally, we examined alternative scenarios in Tables S1 and S2 with SI at 7.5 days with a SD at 3.4 days, which was based on 5 pairs in Li et al.’s study (1).

Full table
Full table
Results
We estimated the time-varying reproduction numbers [R(t)] for the China as a whole, Hubei province and Wuhan respectively. The estimates of R(t) were shown in Figure 2. The estimated transmissibility appeared higher in the early stage of the epidemic. Then, in stage II, the R(t) gradually decreased. By February 15, the reproduction number dropped to 0.72 (95% CI: 0.62–0.82), 0.77 (95% CI: 0.67–0.88) and 0.90 (95% CI: 0.77–1.03) for China, Hubei and Wuhan respectively.
The average reproduction numbers of three stages in China, Hubei and Wuhan are shown in Table 1. In stage I, the reproduction number in China, Hubei province and Wuhan was 4.95 (95% CI: 4.26–5.67), 4.29 (95% CI: 3.66–4.93) and 3.88 (95% CI: 3.30–4.49) respectively. In stage II, the three reproduction numbers dropped to 2.61 (95% CI: 2.47–2.75), 2.76 (95% CI: 2.54–2.95) and 2.71 (95% CI: 2.43–3.01) respectively, indicating that the ‘city lockdown’ of the Wuhan was effective. After the ‘city lockdown’, a series of strict prevention and control measures have been taken nationwide, such as restriction on outdoor activities or gathering, in-home work scheme. Thus, in stage III, the reproduction number decreased further to 0.98, 1.14 and 1.41 respectively.

Full table
Discussion
At present, the reproduction number is estimated at 0.98 (95% CI: 0.96–1.03). In Hubei province, the epidemic situation turns better, with reproduction number 1.14 (95% CI: 1.10–1.19). Although it is still larger than 1 significantly, the downward trend is stable and is likely to be less than 1 shortly in the future. However, the reproduction number in Wuhan is estimated at 1.41 (95% CI: 1.35–1.48) and thus, enhancement in the control measures is supposed to be maintained.
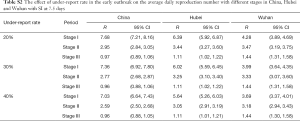
Since January 27, a new coronavirus nucleic acid detection kit has been launched around the country, which greatly improves the detection rate of the disease. However, severe under-reporting phenomena caused by the imbalance regional detection level existed before the appearance of the new coronavirus nucleic acid detection kit (11,22). It is difficult to estimate the specific value of under-reporting rate. According to (16), under-reporting has a very small influence on the reproduction number calculated by this method. In this paper, we assumed that the reporting rate was 20%, 30% and 40% respectively. After correction, we recalculated the average daily reproduction numbers (Table 2) of three stages of China, Hubei and Wuhan.
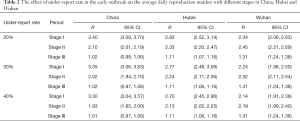
Full table
From Table 2, the average reproduction number of the stage I was lower than that of the original data which was not corrected. Therefore, the actual average reproduction number in the stage I might not be as large as that in Table 1, and the risk of disease had been overestimated. In the stage II and stage III, the rate of under-reporting had little effect on their reproduction numbers.
In order to protect the lives and health of the people, guarantee the prevention and control of the epidemic and ensure transportation of production and living materials, etc., from February 17 to the end of the epidemic prevention and control work, all vehicles are exempt from express away toll according to notice of Ministry of Transport of China (23) to encourage travelling by private cars. Meanwhile, the passenger number of long-distance buses and waterway passenger liners should be controlled under 50% of corresponding passenger load factors and passengers are to be guided to maintain a certain distance (24). This might reduce the possibility of a second outbreak.
In fact, the number of new cases per day in other provinces (exclude Hubei) has declined continually for 13 days, and the epidemic is expected to end in the near future.
The research data in this paper is as of February 16, and in the future, the transmission of the disease is supposed to be considered again and always be considered when calculating a new reproduction number.
Conclusions
We estimated the time-varying reproduction numbers of China, Hubei and Wuhan. The results indicate the improvement and effectiveness of the COVID-19 control measures, although the population in Wuhan is still at a higher risk of infection. As such, we recommend to maintain the enhancement in the public health control efforts in combating the ongoing COVID-19 epidemic.
Acknowledgments
Funding: This work was supported in part by the Natural Science Foundation of China (11961071, 81673275), PhD research startup foundation of Xinjiang Medical University (2018-12), the National S&T Major Project Foundation of China (2018ZX10715002–004–002, 2018ZX10713001–001), program for Tianshan Innovative Research Team of Xinjiang Uygur Autonomous Region, China (2020D14020).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-1944). DH reports grants from Alibaba (China) - HKPU collaborative research project, during the conduct of the study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The data of the confirmed COVID-19 cases were collected via the National Health Commission of the People’s Republic of China, and thus neither ethical approval nor individual consent was not applicable.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Li Q, Guan X, Wu P, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med 2020;382:1199-207. [Crossref] [PubMed]
- Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet 2020;395:689-97. [Crossref] [PubMed]
- Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020;25:2000045. [Crossref] [PubMed]
- Patel A, Jernigan DB. Initial Public Health Response and Interim Clinical Guidance for the 2019 Novel Coronavirus Outbreak - United States, December 31, 2019-February 4, 2020. MMWR Morb Mortal Wkly Rep 2020;69:140-6. [Crossref] [PubMed]
- Carlos WG, Dela Cruz CS, Cao B, et al. Novel Wuhan (2019-nCoV) Coronavirus. Am J Respir Crit Care Med 2020;201:7-8. [Crossref] [PubMed]
- Anderson RM, May RM. Infectious diseases of humans: dynamics and control. Oxford: Oxford University Press, 1991.
- Dietz K. The estimation of the basic reproduction number for infectious diseases. Stat Methods Med Res 1993;2:23-41. [Crossref] [PubMed]
- Diekmann O, Heesterbeek JAP. Mathematical epidemiology of infectious diseases: model building, analysis and interpretation. Int J Epidemiol 2001;30:185-8.
- Eichner M, Dietz K. Transmission potential of smallpox: estimates based on detailed data from an outbreak. Am J Epidemiol 2003;158:110-7. [Crossref] [PubMed]
- Tang B, Wang X, Li Q, et al. Estimation of the Transmission Risk of the 2019-nCoV and Its Implication for Public Health Interventions. J Clin Med 2020;9:E462. [Crossref] [PubMed]
- Zhao S, Musa SS, Lin Q, et al. Estimating the Unreported Number of Novel Coronavirus (2019-nCoV) Cases in China in the First Half of January 2020: A Data-Driven Modelling Analysis of the Early Outbreak. J Clin Med 2020;9:E388. [Crossref] [PubMed]
- Zhao S, Lin Q, Ran J, et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int J Infect Dis 2020;92:214-7. [Crossref] [PubMed]
- Shen M, Peng Z, Xiao Y, et al. Modelling the epidemic trend of the 2019 novel coronavirus outbreak in China. bioRxiv 2020. doi:. [Crossref]
- Riou J, Althaus CL. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveill 2020;25:2000058. [Crossref] [PubMed]
- National Health Commission of the People’s Republic of China, Latest update on outbreak prevention and control. Available online: [Accessed 17 Feb 2020].http://www.nhc.gov.cn/yzygj/s7652m/202002/e84bd30142ab4d8982326326e4db22ea.shtml
- Wallinga J, Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am J Epidemiol 2004;160:509-16. [Crossref] [PubMed]
- Cori A, Ferguson NM, Fraser C, et al. A new framework and software to estimate time-varying reproduction numbers during epidemics. Am J Epidemiol 2013;178:1505-12. [Crossref] [PubMed]
- Ali ST, Kadi AS, Ferguson NM. Transmission dynamics of the 2009 influenza A (H1N1) pandemic in India: the impact of holiday-related school closure. Epidemics 2013;5:157-63. [Crossref] [PubMed]
- Fraser C. Estimating individual and household reproduction numbers in an emerging epidemic. PLoS One 2007;2:e758. [Crossref] [PubMed]
- Fine PEM. The interval between successive cases of an infectious disease. Am J Epidemiol 2003;158:1039-47. [Crossref] [PubMed]
- You C, Deng Y, Hu W, et al. Estimation of the Time-Varying Reproduction Number of COVID-19 Outbreak in China. medRxiv 2020; doi: https://doi.org/. [Crossref]
- Tuite AR, Fisman DN. Reporting, Epidemic Growth, and Reproduction Numbers for the 2019 Novel Coronavirus (2019-nCoV) Epidemic. Ann Intern Med 2020. [Crossref] [PubMed]
- Ministry of Transport of the people’s republic of China, Notice of the Ministry of Transport on Toll Free Highway Vehicle Tolls During the Prevention and Control of New Crown Pneumonia. Available online: [Accessed 18 Feb 2020].http://www.mot.gov.cn/jiaotongyaowen/202002/t20200215_3333512.html
- Ministry of Transport of the People’s Republic of China, Urgent Notice of the Ministry of Transport on ensuring Peak Transport Services for Spring Festival Return. Available online: [Accessed 18 Feb 2020].http://www.mot.gov.cn/jiaotongyaowen/202002/t20200207_3330804.html


