Downregulation of long non-coding RNA LINP1 inhibits the malignant progression of esophageal squamous cell carcinoma
Introduction
Esophageal cancer (EC) has a high mortality rate (more than 300,000 deaths per year) globally and is a very popular malignant tumor worldwide (1). Esophageal squamous cell carcinoma (ESCC) accounts for the majority of ECs in China (2,3). Although the use of chemotherapeutic drugs and targeted drugs has improved the prognosis of patients diagnosed at early stages, EC patients at the progressive stage or those experiencing relapse have a poor prognosis (4,5). The key molecular and genetic mechanisms underlying esophageal carcinogenesis remain unclear. Therefore, improving our knowledge of the molecular mechanisms of ESCC and identifying novel treatment strategies remain urgent needs
Long noncoding RNAs (lncRNAs), which are defined as a group of nonprotein-coding transcripts that are longer than 200 nucleotides (6), are involved in the gene regulation in multiple biological processes (BPs), especially in tumor initiation and progression (7). Increasing evidence supports that lncRNAs have important functions of tumor suppressor and oncogenic pathways at epigenetic, transcriptional, and posttranscriptional levels in many cancers (8-10).
LncRNA in non-homologous end joining pathway 1 (LINP1), located on chromosome 10, is an intergenic lncRNA. LINP1 involves in the progression of various cancers (11-16). However, the role of LINP1 in ESCC is unclear. In the current research, bioinformatics analysis revealed that LINP1 is aberrantly expressed in EC tissues. Analysis of patient tissue samples showed that high LINP1 expression is related with bad prognosis. In vitro functional experiments and in vivo animal experiments demonstrated that LINP1 induces epithelial-mesenchymal transition (EMT) and inhibits apoptosis, thereby promoting the proliferation and metastasis of ESCC cells. These results demonstrate the oncogenic role of LINP1 in ESCC for the first time, and indicate that LINP1 is a candidate therapeutic target for the treatment of ESCC.
Methods
Bioinformatics analysis
The lncRNA chip was selected based on whole transcriptome (WT) expression profiling detection technology produced by Affymetrix (OE WT lncRNA array). The samples of lncRNA array were ten paired of ESCC tissues and normal epithelial tissues collected from ten patients with ESCC. Those ESCC tissues and their corresponding noncancerous mucosal tissues were pathologically verified by immunohistochemistry (Figure S1). The raw data were normalized using the robust multi-array average (RMA) algorithm. Principal component analysis (PCA) was used to analyze the sample data. One-way between-subject ANOVA and fold change were applied to detect differential expression genes between ESCC and normal epithelial tissues. The P value and fold change of all mRNAs and lncRNAs were calculated; the default gene screening criteria were P<0.05 and fold change >2 or <−2. The STRING database (STRING v10.0) was used to analyze differential expression genes in ESCC tissues, and to construct gene network maps. The protein-protein interaction (PPI) network map was analyzed by Cytoscape software. Based on the connectivity of gene interactions in PPI networks, the molecular complex detection (MCODE) algorithm was used to further analyze the PPI network map and screen the optimal model of gene interactions. The gene ontology (GO) network database was used to analyze the differential expression genes in ESCC tissues. The ENRICHR (http://amp.pharm.mssm.edu/Enrichr/) database was used for Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis to identify the biological pathways of enriched protein coding genes and their distribution.
Patient survival
This research was approved by the Research Ethics Committee of Zhongshan Hospital, Fudan University (2018-127) and the informed consent form was obtained from each patient. All cases and tissue specimens were collected from Department of Thoracic Surgery, Zhongshan Hospital, Fudan University. The mRNA were extracted from paired of ESCC tissues and normal epithelial tissues of 122 ESCC patients. The mRNA were reverse transcribed into cDNA, and evaluated by quantitative real-time PCR (qRT-PCR). The results were analyzed by receiver operating characteristic (ROC) curve. A cut-off value was established to distinguish high from low LINP1 expression. Patients with ESCC were divided into LINP1-positive and -negative patients according to the cut-off value. Gender, age, tumor location, tumor size, degree of differentiation, stage, prognosis, survival time, and other characteristics were analyzed.
Fluorescence in situ hybridization assay
LINP1 expression was detected by FISH using the RiboTM Fluorescent In Situ Hybridization Kit (RiboBio, Guangzhou, China), which was patented by State Intellectual Property Office of China in 2016 (Patent No. 201611041669.0). Briefly, 6-µm-thick sections were incubated with FISH Probe Mix for 12 hours at 37 °C. Then the sections were stained with DAPI after washing and observed by confocal microscopy. The lncRNA probe was labeled by Cy3; the emission wavelength was 570 nm and the excitation wavelength was 550 nm. DAPI-labeled nuclei were detected at an excitation wavelength of 364 nm and an emission wavelength of 454 nm.
Cell culture
The EC cell line EC9706 was acquired from American Type Cell Culture (Manassas, VA, USA) and was identified by STR profiling by Hanyin Biotechnology Limited Company (Shanghai). The cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin solution at 37 °C and 5% CO2.
Plasmid construction, cell transfection, and viral infection
The DNA oligonucleotides designed to generate short hairpin (sh)RNAs against the open reading frame of LINP1 mRNA were as follows: 5'-TCTTGATTCAGCTGCATAAAT-3' (LINP1 sh1), 5'-ATGTAACTGATCCTTAGATAT-3' (LINP1 sh2), 5'-GGGATCGGGTTGTTGTTAATT-3' (LINP1 sh3), and 5'-GATAGGAACCCCAGGGAATAA-3' (LINP1 sh4). PHY-310/LINP1 shRNA EC9706 cells were established. The control vector was similarly constructed by directly inserting a scrambled shRNA (Scr sh) into PHY-310. Puromycin was used to select the infected cells.
Reverse transcription PCR and quantitative real-time PCR
Total RNA extraction of EC9706 cells and esophageal carcinoma tissues by using Trizol was performed, and then reverse transcribed to prepare cDNA by Superscript II reverse transcriptase. The target gene was amplified using the FastStart universal SYBR Green Master (ROX) kit by the mothed of qRT-PCR. The primers used for qRT-PCR were as follows: LINP1-forward (F): 5'-CTTGACTCTGGGTGGGCTGTGT-3'; LINP1-reverse (R): 5'-AGGTGGCTCTGTTCTGGGTGAC-3'; E-cadherin-F: 5'-CGAGAGCTACACGTTCACGG-3'; E-cadherin-R: 5'-GGGTGTCGAGGGAAAAATAGG-3'; N-cadherin-F: 5'-AGCCAACCTTAACTGAGGAGT-3'; N-cadherin-R: 5'-GGCAAGTTGATTGGAGGGATG-3'; Vimentin-F: 5'-GACGCCATCAACACCGAGTT-3'; Vimentin-R: 5'-CTTTGTCGTTGGTTAGCTGGT-3'; Snail-F: 5'-TCGGAAGCCTAACTACAGCGA-3'; Snail-R: 5'-AGATGAGCATTGGCAGCGAG-3'; Bcl-2-F: 5'-GGTGGGGTCATGTGTGTGG-3'; Bcl-2-R: 5'-CGGTTCAGGTACTCAGTCATCC-3'; Bax-F: 5'-CCCGAGAGGTCTTTTTCCGAG-3'; Bax-R: 5'-CCAGCCCATGATGGTTCTGAT-3'; CDK1-F: 5'-AAACTACAGGTCAAGTGGTAGCC-3'; CDK1-R: 5'-TCCTGCATAAGCACATCCTGA-3'; β-actin-F: 5'-CCTGGCACCCAGCACAAT-3'; β-actin-R: 5'-GCTGATCCACATCTGCTGGAA-3'.
Cell proliferation assay
Cells were seeded at a density of 1.5×103 cells per well in 96-well culture plates (Corning Inc., Corning, NY, USA) in 100 µL medium and cultured for 7 days. Cell growth was detected by adding 10 µL Alamar Blue (Yeasen Biotech Co. Ltd, CAT: 40202ES92) into each well.
Colony formation assay
A total of 1000 cells were mixed with 2 mL of medium and then placed onto a 6-well plate in triplicate. After 10 days of incubation, the cells were washed twice with phosphate buffer saline (PBS). Then the cells were fixed with methanol for 15 min and then stained with 0.1% crystal violet for 20 min, and then photographed and counted.
Wound healing assay
Cells (5×105) were grown overnight in a 6-well culture dish. The cell monolayer was scratched with a pipette tip and photographed at 0 and 48 h. The scratched area was measured in a 100× magnified visual field. The migration index was calculated to evaluate the cell migration capacity. Experiments were conducted in triplicate.
Transwell migration assay
Cell migration capacity was analyzed using a Transwell chamber with an 8-µm (pore size) polycarbonate filter (BD Biosciences, USA). Briefly, 2.5×104 cells in 100 µL serum-free DMEM were placed onto the upper chambers. The lower chambers were loaded with 600 µL medium containing 10% FBS. After incubation for 48 h at 37 °C, the invaded cells were fixed with methanol for 1 h, stained with 0.1% crystal violet for 10 min, and photographed under a light microscope (×100).
Flow cytometry
Cell cycle was evaluated by the BD cycle detection kit. Briefly, cells at the logarithmic growth phase were digested with pancreatic enzymes without EDTA. A total of 5×105 cells were fixed with 2 mL 75% ethanol for 2 hour at 4 °C and then stained with 0.5 mL PI solution (BD Pharmingen, Cat: 550825), followed by incubation in the dark for half an hour. Flow cytometry was performed using the FACSCalibur flow cytometer (BD Pharmingen), and the results were analyzed using ModFit cell cycle software.
Apoptosis was evaluated using the Annexin V-PE/7-AAD double-stained cell apoptosis assay kit (C1065L Beyotime, China); 1×105 cells were resuspended with mixture consisting of 195 µL of binding buffer and 5 µL of Annexin V-PE and 7-AAD. The cell suspension was incubated for 15 min at room temperature. Flow cytometry was performed, and the results were analyzed using FlowJo 7.6 software.
Western blot assay
Antibodies against the following proteins were from Affinity, including E-cadherin (AF0131), N-cadherin (AF4039), vimentin (AF7013), SNAIL (AF6032), Bax (AF0120), Bcl-2 (AF6139), CDK1 (AF6024), and β-actin antibody from Abcam (ab8227). For western blot assays, the membrane was incubated with primary antibodies overnight at 4 °C. The secondary antibodies were goat anti-mouse IgG (Beyotime, A0216) or goat anti-rabbit IgG (Beyotime, A0208). The ECL chemiluminescence method was used to quantify the gray values of bands using Image J software.
Xenograft tumor assay
All animals have received humane care in compliance with the 1996 “Guide for the Care and Use of Laboratory Animals” as recommended by the US National Institutes of Health (NIH). Animal experiments were approved by the Research Ethics Committee of Zhongshan Hospital, Fudan University and conducted in accordance with institutional guidelines and protocols. Seventy-five mice were divided into three groups randomly (n=25). EC9706 negative control (NC), shRNA-scr, and shRNA-LINP1 cells (5×106) were injected into the skin of 6-week-old female BALB/C nude mice. The subcutaneous tumor was assessed by a Vernier caliper every 3 days. The longest diameter “a” and the shortest diameter “b” of the tumor were measured, and the tumor volume was calculated using the following formula: v (unit: mm3) = a × b2 ×0.5. Mice were sacrificed by day 21, weighed, and tumors were measured.
Data analysis
Data were analyzed using the SPSS 24.0 software (SPSS Inc., USA). Differences between multiple sets of numerical variables were determined using analysis of variance (ANOVA), and differences between two sets of continuous variables were analyzed using the t-test. The chi-square test was used for categorical variables. The ROC curve was constructed using a nonparametric method, and the independent prognostic factors affecting prognosis were identified by Kaplan-Meier analyses and Cox regression multivariate analyses. Two-tailed P<0.05 was considered statistically significant.
Results
Bioinformatics analysis
The lncRNA chip scanning image showed high quality signals (Figure S2). After log conversion of the lncRNA chip CHP file, the chip data distribution of ESCC and normal epithelial tissue was consistent.
The intensity of the probe unit was concentrated in the range of 5.79–8.97, and the signal data were concentrated to 2.82–9.19, and no abnormally distributed data were detected (Figure S3A,B). Box plot analysis showed the relative signal data of lncRNA chips in all tissues were basically consistent, with no abnormal outliers (Figure S3C). The above results showed that the chip data had high stability.

Pearson correlation and Spearman correlation analyses showed a high correlation between normal tissues and normal tissues, as well as between ESCC tissues and ESCC tissues, and a low correlation between normal epithelial tissues and ESCC (Figure S3D,E). PCA results revealed that the distribution of lncRNA microarray data between normal epithelial tissue and ESCC tissue had obvious boundaries (Figure S3F).
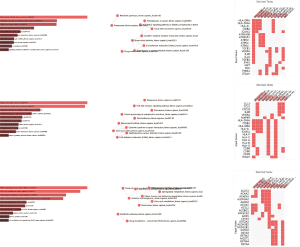
A total of 90,675 genes were detected in the two groups, including 27,133 protein-coding genes and 63,542 lncRNAs. There were 5,621 differentially expressed genes between ESCC and normal epithelial tissues, including 3,185 protein-coding genes and 2,436 lncRNAs. Of these, 3,248 genes were upregulated in ESCC, including 1,939 protein-coding genes and 1,309 lncRNAs, whereas 2,373 genes were downregulated in ESCC, including 1,246 protein-coding genes and 1,127 lncRNAs (Figure 1A). According to the mRNA and lncRNA expression levels in different tissues, we performed hierarchical clustering analysis of the chip data and generated a heatmap to directly visualize the differences in mRNA and lncRNA expression profiles between ESCC and normal esophageal tissues (Figure 1B). LINP1 (fold change =556.2, P<0.001) was selected for further analysis because it showed the most significant difference in expression among lncRNAs. We randomly selected 12 significant differentially expressed genes from the microarray, and their expression was validated by qRT-PCR (Figure 1C). The results revealed no significant difference, indicating a favorable consistency.
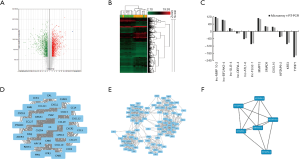
GO includes three structural ontologies to analyze the functional properties of genes in a species-specific manner, namely, BP, cellular component (CC), and molecular function (MF). The top ten significantly enriched GO terms of BP, CC, and MF for all up- and down-regulated genes based on the P value are shown in Tables S1-S3.
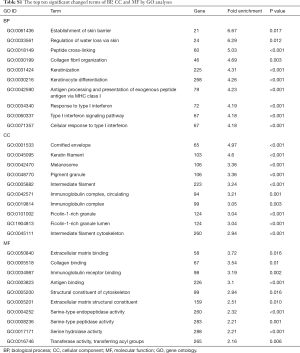
Full table
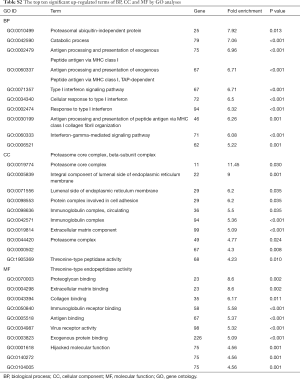
Full table
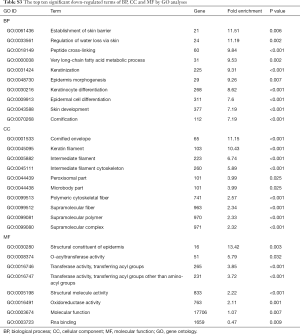
Full table
KEGG analysis showed that differential expression genes in ESCC tissues were mainly concentrated in 31 biological pathways. The top five significantly enriched KEGG pathways were rheumatoid arthritis, staphylococcus aureus infection, phagosome, metabolic pathways and focal adhesion (Figure S4A,B,C). The top five KEGG pathways containing the largest quantity of enriched genes were metabolic pathways, pathways in cancer, cytokine-cytokine receptor interaction, phagosome, and focal adhesion. The top ten significantly enriched up- and down-regulated KEGG pathways are listed in Figure S4D,E,F and Figure S4G,H,I.
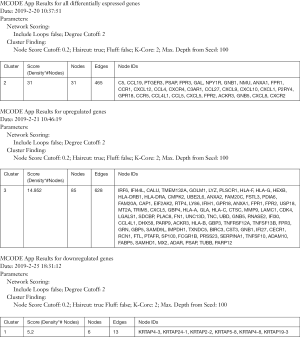
The MCODE algorithm was applied to analyze the optimal PPI network map. The PPI network map showed the optimal interaction models (score =31, nodes =31, edges =465, Figure 1D) of differentially expressed genes. We also constructed a protein network interaction model of upregulated genes (score =14.952, nodes =85, edges =628, Figure 1E), and downregulated genes (score =5.2, nodes =6, edges =13, Figure 1F). The list of identified candidate genes were shown in Supplement I.
High expression of LINP1 in ESCC tissues indicates poor prognosis
Those ESCC tissues and their corresponding noncancerous mucosal tissues was pathologically verified by immunohistochemistry (Figure S1). LINP1 expression level was evaluated by qRT-PCR in 122 ESCC and equal numbers of normal esophageal tissues, showing that ESCC had higher LINP1 expression than normal esophageal tissues (17.656 vs. 2.53, P<0.001; Figure 2A). The ROC curve was constructed using a nonparametric method, and the area under the ROC curve was 0.819 [95% confidence interval (CI): 0.763–0.867]. The calculated cut-off value was 9.135 (Figure 2B). The expression of LINP1 was analyzed in 122 ESCC tissues by qRT-PCR. ESCC patients were separated into a LINP1 high expression group (n=71) and a LINP1 low expression group (n=51) according to the cut-off value. The LINP1 high expression group had a larger tumor size (P=0.009), more local invasion (P=0.015), higher lymph nodes (LN) stage (P=0.044), and more advanced TNM stage (P=0.010), whereas no significant differences in age, gender, tumor location, and tumor differentiation were observed (Table 1).

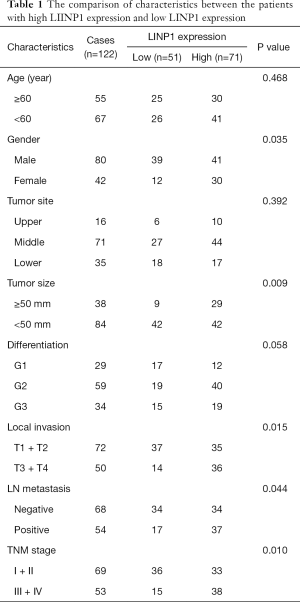
Full table
Univariate and multivariate analyses identified tumor differentiation, local invasion, LN metastasis, TNM stage, and LINP1 expression as independent prognostic factors (Table 2). Patients with high LINP1 expression were associated with worse survival (hazard ratio =1.445, 95% CI: 1.094–2.310; Figure 2C).
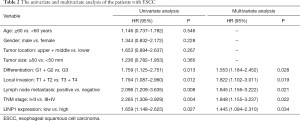
Full table
LINP1 highly expressed in ESCC tissues and cells
FISH assay showed that LINP1 was expressed at low levels in normal esophageal tissues (Figure 2D), whereas it was highly expressed in ESCC and mainly expressed in the cytoplasm (Figure 2E). The results of FISH assays in EC9706 cell lines were consistent with those obtained in tissues, and LINP1 was mainly expressed in the cytoplasm (Figure 2F).
LINP1 downregulation inhibits the proliferation and migration of EC9706 cells
To explore the role of LINP1 in human EC, LINP1 sh1, sh2, sh3, and sh4 EC9706 cell lines were generated and compared with the corresponding control cell lines (shRNA-scr). qRT-PCR analysis showed that LINP1 mRNA was significantly decreased in sh1 (40.26%), sh2 (61.75%), sh3 (54.55%), and sh4 (60.20%) cells compared with control cells (all P<0.001; Figure 3A). The LINP1 sh2 cell line was selected for further experiments because it showed the most significant knockdown effect. The proliferation rate of shRNA-LINP1 cells was significantly lower than that of the NC and shRNA-scr cells (P<0.001; Figure 3B). Consistent with the proliferation results, the colony formation assay revealed that shRNA-LINP1 cells formed significantly fewer colonies than NC and shRNA-scr cells (P<0.001; Figure 3C,D).
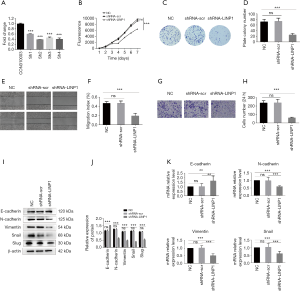
Scratch test indicated that the migration rate of shRNA-LINP1 cells was slower than that of the control cell lines (migration index, 46.4% vs. 46.6% vs. 19.1%; P<0.001; Figure 3E,F). Transwell migration assays showed that knockdown of LINP1 significantly reduced EC9706 cell migration (P<0.001; Figure 3G,H). Western blot analysis of EMT markers showed that knockdown of LINP1 significantly downregulated the expression of N-cadherin, vimentin, snail and slug, and upregulated the expression of E-cadherin in EC9706 cells compared with that in the two control cell lines (all P<0.001; Figure 3I,J). qRT-PCR analysis showed that the mRNA expression of N-cadherin, vimentin, and snail significantly reduced by 40.0%, 48.8%, and 39.4%, whereas the expression of E-cadherin increased by 63.6% compared with that in NC cells (all P<0.01; Figure 3K). Taken together, LINP1 knockdown inhibited the proliferation and migration of EC9706 cells by modulating the EMT process.
Knockdown of LINP1 affects the cell cycle and promotes apoptosis
Flow cytometric analysis showed a significant increase in the proportion of G2/M phase cells in the shRNA-LINP1 group compared with that in NC and shRNA-scr cells (10.92% vs. 10.99% vs. 18.25%, P<0.001), concomitant with a significant decrease in the number of cells in S phase (28.78% vs. 28.58% vs. 21.35%, P<0.001). Therefore, LINP1 knockdown promoted cell cycle arrest in the G2/M phase (Figure 4A,B). The mean rate of apoptosis in NC and shRNA-scr cells was 4.40% and 4.24%, respectively, whereas that in shRNA-LINP1 cells was 13.68% (P<0.001), which suggested that LINP1 knockdown promoted EC9706 cells apoptosis (Figure 4C,D).
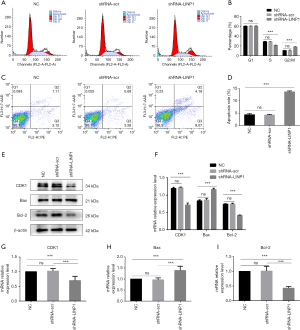
To explore the possible mechanism of LINP1 on cell cycle and apoptosis in EC9706 cells, the expression of related markers was assessed by qRT-PCR and western blotting. LINP1 knockdown significantly downregulated the mRNA and protein expression of CDK1 (P<0.001). Assessment of apoptosis markers showed that Bcl-2 was decreased (P<0.001) whereas Bax was increased (P<0.001) (Figure 4E,F,G,H,I).
Knockdown of LINP1 suppresses EC tumorigenesis in vivo
To determine whether LINP1 contributes to the development of EC in vivo, we performed animal experiments by generating xenograft tumors through injection of NC, shRNA-scr, and shRNA-LINP1 EC9706 cells (Figure 5A). The tumor growth rate of the shRNA-LINP1 cell injected group was significantly slower than that of the control groups (P<0.001; Figure 5B), and the tumors in shRNA-LINP1 group have lower weight than those in the control groups (1.04 vs. 3.39 vs. 3.06 g, P<0.001; Figure 5C). The tumor inhibitory rate was 69.3% after knockdown of LINP1 in EC9706 cells. These observations suggested that LINP1 knockdown exerted significant inhibitory effects in ESCC cells.

Discussion
Many lncRNAs are associated with cancer prognosis (17-22). However, the diagnosis and treatment for ESCC requires effective predictive biomarkers. This study showed that the most significant differentially expressed lncRNA was LINP1. LINP1 was first identified as a novel lncRNA by Zhang et al. (11) in breast cancer. LINP1 participate in the development of breast cancer and associated with poor prognosis (11-13). The involvement of LINP1 in other cancers was also reported (14-16). However, no studies so far have been done in the role of LINP1 in ESCC. This research examined the biological function of LINP1 in ESCC in vitro and in vivo.
We analyzed the correlation between LINP1 expression and the clinical characteristics in ESCC patients, and found that high expression of LINP1 was significantly correlated with advanced tumor stage, suggesting LINP1 played a carcinogenic role in ESCC. We also revealed that patients with high LINP1 levels were associated with poorer prognoses compared with those with low LINP1 levels (P=0.027). Liang et al. (12) also demonstrated that increased LINP1 expression levels were significantly correlated with distant metastasis and advanced clinical stage in breast cancer, and associated with an unfavorable outcome in breast cancer patients. Wu and associates (15) suggested that high LINP1 expression is related with higher tumor stage and worse prognosis. However, these studies did not provide a basis for determining the level of gene expression. In the present study, we used ROC curve analysis to determine a cut-off value of LINP1 expression, which helped avoid subjectivity and improved the accuracy of clinical judgement.
This study showed that LINP1 knockdown inhibited the proliferation and migration of ESCC cells possibly by causing cell cycle arrest at the G2/M phase. CDK1 downregulation, which was confirmed by western blotting, indicated that LINP1 may regulate cell cycle progression by modulating CDK1 expression. This was not consistent with the results of Liang et al., who reported that LINP1 knockdown inhibited cell proliferation by inducing G1-phase cell cycle arrest (12). The different results may be due to the different genetic backgrounds of cancer cells. In addition, we demonstrated that LINP1 knockdown significantly increased the apoptotic rate, suggesting that knockdown of LINP1 promoted apoptosis of EC9706 cells. Consistently, LINP1 knockdown significantly downregulated Bcl-2 and upregulated Bax, as shown by western blotting. Wang et al. (14) demonstrated that LINP1 downregulation increases irradiation-induced cell apoptosis in cervical cancer. EMT is a crucial process for early metastasis from primary tumors. In this process, epithelial-derived cancer cells lose the characteristics of epithelial cells and acquire mesenchymal properties, conferring cells migration, invasion, and stem cell capabilities (23). We found that downregulation of LINP1 suppressed the EMT procession by affecting key regulatory factors, including upregulation of E-cadherin and downregulation of N-cadherin, vimentin, snail and slug. Liang and coworkers (12) showed similar results in breast cancer. These findings suggest that LINP1 exerts a vital role in the ESCC development.
We provide evidence that LINP1 has unrecognized roles in carcinogenesis and development of ESCC. However, this study had several limitations. Firstly, only a few pairs of microarray specimens were tested. Secondly, the number of ESCC specimens was small, which may lead to selection bias. Thirdly, a single cell line was used in which the target gene was downregulated for cell-based experiments. Multiple ESCC cell lines and stable cell line overexpressing LINP1 should be used to clarify the biological roles of LINP1 in malignant behavior of ESCC cells. More human esophageal squamous cell lines needed to verify the results. In addition, we did not construct ESCC cells with LINP1 overexpression. We intend to overcome the flaws of this study and perform overexpression experiments and further verification in our future studies. Also, we will carry out the follow-up experiments to verify the results of pro-apoptotic factor, such as FasL, TNFa and TRAIL, and perform the immunohistochemical staining for the apoptosis-related protein expression in the xenotransplanted tumors of mice. Treatment modalities have significant prognostic value, such as radio- or chemo-therapy, neo-adjuvant or adjuvant treatment. But we did not discuss the efficacy of treatment modalities, because treatments were various and the chemotherapy regimens as well as radiotherapy doses were inconsistent. Further randomized clinical trial would clarify the benefit of ESCC treatment. Finally, research into the underlying molecular mechanism was insufficient, and the specific binding of miRNA or RNA and the sites in LINP1 were not identified. Further study is needed to explore the function of LINP1 in ESCC. Although our data showed the potency of LINP1 silencing in inhibition of cell proliferation and induction of apoptosis, more evidence are needed to clarify if LINP1 is a driver gene in ESCC. Lentiviral-based shRNA might be utilized as an effective cancer therapy based on previous study (24). Additionally, the blockage of tumorigenesis by lentiviral-mediated LINP1 shRNA also supported the effectiveness of this strategy in ESCC. However, it is still premature to conclude LINP1 is a targetable gene. It is necessary to justify if LINP1 is a therapeutic target in ESCC by animal experiments and clinical trials.
Supplement I
Acknowledgments
Funding: The study was supported by Shanghai Sailing Program (No. 17YF1402400).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-1009). SW serves as an unpaid Section Editor of Annals of Translational Medicine from Oct 2019 to Sep 2020. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This research was approved by the Research Ethics Committee of Zhongshan Hospital, Fudan University (2018-127) and the informed consent form was obtained from each patient. All animals have received humane care in compliance with the 1996 “Guide for the Care and Use of Laboratory Animals” as recommended by the US National Institutes of Health (NIH). Animal experiments were approved by the Research Ethics Committee of Zhongshan Hospital, Fudan University and conducted in accordance with institutional guidelines and protocols.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Holmes RS, Vaughan TL. Epidemiology and pathogenesis of esophageal cancer. Semin Radiat Oncol 2007;17:2-9. [Crossref] [PubMed]
- Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med 2003;349:2241-52. [Crossref] [PubMed]
- Samson P, Puri V, Lockhart AC, et al. Adjuvant chemotherapy for patients with pathologic node-positive esophageal cancer after induction chemotherapy is associated with improved survival. J Thorac Cardiovasc Surg 2018;156:1725-35. [Crossref] [PubMed]
- Digklia A, Voutsadakis IA. Targeted treatments for metastatic esophageal squamous cell cancer. World J Gastrointest Oncol 2013;5:88-96. [Crossref] [PubMed]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013;154:26-46. [Crossref] [PubMed]
- Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017;36:5661-7. [Crossref] [PubMed]
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6. [Crossref] [PubMed]
- Lin C, Wang Y, Wang Y, et al. Transcriptional and posttranscriptional regulation of HOXA13 by lncRNA HOTTIP facilitates tumorigenesis and metastasis in esophageal squamous carcinoma cells. Oncogene 2017;36:5392-406. [Crossref] [PubMed]
- Lin A, Li C, Xing Z, et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol 2016;18:213-24. [Crossref] [PubMed]
- Zhang Y, He Q, Hu Z, et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat Struct Mol Biol 2016;23:522-30. [Crossref] [PubMed]
- Liang Y, Li Y, Song X, et al. Long noncoding RNA LINP1 acts as an oncogene and promotes chemoresistance in breast cancer. Cancer Biol Ther 2018;19:120-31. [Crossref] [PubMed]
- Liu XM, Yang B, Han J. Increased long noncoding RNA LINP1 expression and its prognostic significance in human breast cancer. Eur Rev Med Pharmacol Sci 2018;22:8749-54. [PubMed]
- Wang X, Liu H, Shi L, et al. LINP1 facilitates DNA damage repair through non-homologous end joining (NHEJ) pathway and subsequently decreases the sensitivity of cervical cancer cells to ionizing radiation. Cell Cycle 2018;17:439-47. [Crossref] [PubMed]
- Wu HF, Ren LG, Xiao JQ, et al. Long non-coding RNA LINP1 promotes the malignant progression of prostate cancer by regulating p53. Eur Rev Med Pharmacol Sci 2018;22:4467-76. [PubMed]
- Zhang C, Hao Y, Wang Y, et al. TGF-beta/SMAD4-Regulated LncRNA-LINP1 Inhibits Epithelial-Mesenchymal Transition in Lung Cancer. Int J Biol Sci 2018;14:1715-23. [Crossref] [PubMed]
- Zhang Y, Jin X, Wang Z, et al. Downregulation of SNHG1 suppresses cell proliferation and invasion by regulating Notch signaling pathway in esophageal squamous cell cancer. Cancer Biomark 2017;21:89-96. [Crossref] [PubMed]
- Zang W, Wang T, Huang J, et al. Long noncoding RNA PEG10 regulates proliferation and invasion of esophageal cancer cells. Cancer Gene Ther 2015;22:138-44. [Crossref] [PubMed]
- Chen X, Kong J, Ma Z, et al. Up regulation of the long non-coding RNA NEAT1 promotes esophageal squamous cell carcinoma cell progression and correlates with poor prognosis. Am J Cancer Res 2015;5:2808-15. [PubMed]
- Li X, Wu Z, Mei Q, et al. Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br J Cancer 2013;109:2266-78. [Crossref] [PubMed]
- Lin C, Zhang S, Wang Y, et al. Functional Role of a Novel Long Noncoding RNA TTN-AS1 in Esophageal Squamous Cell Carcinoma Progression and Metastasis. Clin Cancer Res 2018;24:486-98. [Crossref] [PubMed]
- Hu L, Wu Y, Tan D, et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res 2015;34:7. [Crossref] [PubMed]
- Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871-90. [Crossref] [PubMed]
- Mishra DK, Balekar N, Mishra PK. Nanoengineered strategies for siRNA delivery: from target assessment to cancer therapeutic efficacy. Drug Deliv Transl Res 2017;7:346-58. [Crossref] [PubMed]