
Vof16-miR-205-Gnb3 axis regulates hippocampal neuron functions in cognitively impaired diabetic rats
Introduction
Diabetes mellitus (DM) is a chronic metabolic disease induced by a deficiency of insulin secretion and/or malfunction. In recent years, the incidence of diabetes has increased and there are 425 million patients suffering from the disease in China. Long-term high blood glucose and blood glucose excursion will damage the kidney, retina and cerebrovascular, cardiovascular, and nervous systems (1).
The association between hyperglycemia and adverse brain outcomes is well established, especially in childhood-onset type 1 diabetes which increases the risk of cognitive impairment with ageing (2). Long-term type 1 diabetes is also associated with impaired immediate and delayed memory. The hippocampus critically contributes to cognition and emotion, and as learning and memory functions are processed within hippocampal circuits and regulated by synaptic plasticity, conditions deleterious to neuroplasticity such as diabetes would impair cognition ability (3-5). Diabetes-induced cognitive decline is related to insulin resistance, which reduces the activation of Akt and increase subsequent GSK3β activity (6,7). The activation of GSK3β phosphorylates tau, whose hyperphosphorylation induces neurofibrillary tangles (8). Insulin resistance-related hyperinsulinemia also impairs the clearance of β-amyloid protein. According to MRI studies, older diabetic patients have a smaller hippocampus, which is probably due to the impairment of neuroprogenitor proliferation.
As a result of the Encyclopedia of DNA Elements project (ECODE), it has been proved that Non-coding RNA (ncRNA) is involved in many biological processes regulation of physical function. ncRNAs, which comprise 98.5% of the human transcriptome, have been proven to be involved in numerous biological and pathophysiological processes (9). MicroRNAs, a class of ~22 nt long non-coding RNAs, are post-transcriptional regulators through transcripts degradation and/or translation inhibition. Long non-coding RNA (lncRNA) is a class of non-protein coding RNA over 200 nucleotide long. Currently, 30,369 lncRNA transcripts have been reported (10) and have been seen to participate in the regulation of adjacent protein-coding (11). A mechanism recently proposed suggests lncRNA could regulate miRNAs negatively when lncRNA share the same miRNA response elements (MREs) with miRNAs. Therefore, lncRNAs act as competing endogenous RNAs (ceRNAs) by competitively binding miRNAs, and derepressing miRNAs subsequently targets gene expression (12). Since both lncRNA and miRNA contribute to RNA metabolism, the aberrant expressions of lncRNAs and/or miRNAs have been proposed as essential players in several human diseases including cancers and metabolic conditions, and this hypothesis is supported by several findings. FAL1-miR-1236 promotes cell proliferation and migration in hepatocellular carcinoma cells (13), LncRNA CDKN2BAS promotes metastasis via the miR-153-5p-ARHGAP18 axis in hepatocellular carcinoma (14), and the HCP5-miR-29b-3p-MMP9/ITGB1 axis was linked to the prognosis of pancreatic cancer (15). However, few studies have addressed lncRNA-miRNA-mRNA regulation in the etiology and development of diabetes encephalopathy.
In this study we analyzed the profiles of lncRNA and mRNA in the hippocampi of STZ-induced diabetic rats. Among the differential expressed lncRNAs and mRNAs, we found that Vof-16, an upregulated lncRNA in STZ-induced diabetic hippocampi, could regulate Gnb3 through sponging miR-205. Furthermore, cell counting assay revealed that Vof-16 regulated hippocampal neuron proliferation partially via miR-205. Altogether, the proposed Vof-16-miR-205-Gnb3 axis might be helpful for the understanding of molecular mechanisms underlying the pathogenesis of STZ-induced diabetic encephalopathy. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-21-2016).
Methods
Animals and induction of type 1 diabetes
Sixty male SD rats (190–240 g, 6 months old) used in this study were purchased from the Department of Laboratory Animal Science of Fudan University, Shanghai. Experiments were performed under a project license (No.: 2018-37) granted by institutional ethics board of Shanghai Tongren Hospital, in compliance with the National Institutes of Health Guide for Care and Use of Laboratory Animals. After 2 weeks of adaptive feeding, the random list randomly divided the selected SD rats into blank control group (CTR group) with 30 rats, and only fed normal diet (containing 14.42% fat, 41.47% carbohydrate and 21.06% protein); 30 rats in the experimental group (STZ group). During the experiment, four rats died in the STZ group and one in the CTR group.
Preparation of diabetic rat model: rats in STZ group were given STZ citrate buffer (55mg/kg) intraperitoneally after 12 hours of fasting and water deprivation. Rats in CTR group were given equal volume of sodium citrate buffer intraperitoneally. Three days later, the tail vein blood of rats was taken to detect the random blood glucose, the blood glucose was more than 16.7 mmol/L, and the fasting blood glucose remained above 16.7 mmol/L for three consecutive weeks. Blood glucose was monitored regularly, and the rats with blood glucose <16.7 mmol/L were excluded according to the blood glucose detection. After the blood glucose was measured, erythromycin ointment was applied to the wound.
HE staining
Hippocampal morphology was detected with HE staining. The prepared paraffin slices were dewaxed and rehydrated using graded ethanol at room temperature and following immersion in drilled water for 7 min, the sections were stained by hematoxylin solution and eosin solution successively according to the standard procedure. The sections were then sealed by neutral balsam, then observed under a microscope.
Y maze
To assess spatial and working memory, the Y-maze test was performed. The maze is a Y shaped apparatus consisting of three isometric arms 30 cm in length, 7 cm in width, and 15 cm in height. Each rat was initially placed at the end of one arm, and after a 5–8-min adaption, the rats were allowed to freely explore the maze. Consecutive entry into three arms in an alternative order was defined as a successive entry and the total reaction time (TRT) and errors number (EN) were recorded.
Morris water maze
To test spatial learning and memory, the Morris water maze was used. This maze was made of a plastic circular tank 15 cm in height and 10 cm in diameter, which was divided into four equal quadrants clockwise.
RNA isolation and quantitative real-time polymerase chain reaction
Total RNA was isolated with Trizol reagent (Invitrogen) and purified with a QIAGEN RNeasy® Kit according to the manufacture’s instruction. The cDNA was synthesized with miScript II RT Kit (QIAGEN) and real time PCR reactions were performed using a miScript SYBR Green PCR Kit (QIAGEN) and conducted on a ViiA™ 7 Real-Time PCR System (ABI). The primers used in this study were as follows (5'-3'): GAPDH: F: CCTGCACCACCAACTGCTTA, R: CATCACGCCACAGCTTTCCA; Vof16: F: ATGGCAGACATTCGGTGCTT, R: AGCATTTGTATGGCGTTGCC, Gnb3: F: CACGTCCGTAGCCTTCTCAC, R: AAGAACGCCTACACGCTCAC.
The cycling program was as follows: the initial denaturation phase, 95 °C for 15 min, followed by the 40 cycles of amplification phase, 94 °C for 15 s, 60 °C for 30 s, and 70 °C for 30 s. The 2−ΔΔCT method was used to normalize the fold change in gene expressions and GAPDH was used as control.
Microarray
To detect the differential expressed lncRNA in hippocampi between diabetic rats and control groups, cRNA was synthesized and amplified from total RNA using the Agilent Low Input Quick Amp Labeling Kit according to manufacturer’s instruction. The Cy3-labeled cRNA was purified with a QIAGEN RNeasy® Kit and the products were hybridized to Agilent Rat lncRNA V3 (8*60K), followed by washing. The microarrays were then scanned using an Agilent DNA microarray scanner. The differential expressed lncRNAs were analyzed as described previously.
Primary rat hippocampal neuron culture and immunofluorescent staining
Before the isolation of hippocampi from brains, the neonatal rats (<24 h) were soaked with 75% ethanol for 1–2 min. The hippocampus was then washed and minced in pre-cold D-hanks, and the tissues were digested with 0.25% trypsin supplied with 0.03% EDTA in a 50-mL tube at 37 °C for 20 min. After a 15-min-incubation, the digestion was stopped with the addition of 5% fetal bovine serum (FBS) (Gibco, USA) and the neurons were then centrifuged and suspended in Neurobasal (Gibco) supplied with 10% FBS and filtered with a 74-µM mesh to remove the debris. The suspensions were centrifuged using lymphocyte separation liquid at 2,000 rpm for 20 min. The supernatant was then discarded, and the cells were resuspended with D-hanks. The neuronal suspensions were plated on coverslips coated with 0.01% poly-L-lysine (Sigma, USA) and cultured in Neurobasal containing 10% FBS in a 5% CO2 incubator at 37 °C. The medium was half replaced every 3–4.5 days.
To identify the isolated primary rat hippocampal neurons, the cells were cultured for 3 days and then fixed with 4% paraformaldehyde for 20 min at room temperature. After being washed three times, the cells were penetrated with 0.1% TritonX-100 (in PBS) for 10 min, followed by blocking with 5% goat serum for 20 min at room temperature. The cells were then incubated with primary antibody mouse anti-Nestin (1:1,000) at 4 °C overnight, followed by 1 h incubation of Cy3-conjugated secondary mouse anti rat NeuN monoclonal antibody (1:500) was incubated at 37 °C for 2 hours and then incubated at 4 °C overnight; biotin labeled Sheep anti mouse IgG (1:200) and avidin anti biotin horseradish peroxidase complex (1:200) were incubated at 37 °C for 2 hours respectively, and each step was 0.01 mmol /L PBS was rinsed for 3 times, and the color was developed by TMB-ST method: distilled water was washed for 3 times; the staining solution was pre incubated at room temperature in dark for 15–20 min; 0.3% H2O2 (3.5 µL/mL) was added for 3 times, with an interval of 10 min each time, and the color was controlled under microscope until the positive cells showed clear green; 0.05 m PBS was added into the sections wash for 5 min × 3 times; mount, dry and seal with neutral gum. The images were obtained with Laser scanning confocal microscope (Olympus Fluo View FV1000).
Cell transfection
For the transfection of 293T cells, cells were seeded in a 24-well plate at a density of 1×105 cells/well. Transfection was performed using Turbofect reagent in accordance with the manufacturer’s instructions.
Cell proliferation detection
Cell proliferation was assessed by a Cell Counting Kit-8 (Dojindo, Japan) according to the manufacturer’s instructions. In brief, the primary cultured hippocampal neurons were seeded into a 96-well plate at a density of 5,000 cells/well. The optical density was then determined with a microplate reader at a wavelength of 450 nm.
Western blotting
Total cell proteins were isolated with RIPA lysis buffer. Proteins were separated by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to PVDF membrane. Membranes were blocked in 5% skim milk in TBST for 1 h at room temperature followed by incubation with primary antibody at 4 °C overnight with Gnb3 antibody or β-actin. The membrane was then washed for 10 minutes three times with TBST followed by incubation with appropriate secondary antibodies for 1 h at room temperature. The blots were visualized with ECL solution and exposed to X-ray film (Kodak) with β-actin used as the control.
Plasmid construction
For lncRNA Vof-16 overexpression, the cDNA of Vof-16 was amplified with PCR and cloned into the psiCHECK vector (Promega) and for miR-205 overexpression, premiR-205 was amplified with PCR and cloned into the PLV. X-PURO vector. For the luciferase reporter assay, wild type or mutant 3'-UTR of Gnb3 containing miR-205 binding site were amplified and cloned into pGL3-Basic.
Dual luciferase assay
HEK-293T cells were seeded in 24-well plates at a density of 1×105 cells/well. After 24 h, the cells were transiently transfected with a mixture of 1.2 µg of DNA and after 48 h, luciferase activity was measured using the dual-luciferase reporter assay system (Promega) according to the manufacture’s instruction.
Statistical analysis
The data were analyzed with SPSS 20.0 (IBM Corp.) and presented as mean ± SD. Comparisons between two groups were made using t test, while comparisons between multiple groups were carried with one-way ANOVA. If the quality characteristic value was not in the Gauss distribution, Kruskal-Wallis analysis was carried out. P<0.05 was indicative of statistically significant difference.
Results
Induction of a diabetic rat model by STZ injection
Diabetes was induced by injection of streptozotocin (STZ). The body weight of the STZ-treated group (N=26) was similar to that in the control group (N=29) at 1 week after treatment (220.1±12.0 vs. 221.0±13.1, P=0.808). However, the body weights were significantly higher in the STZ-treated group than in the control group at 9 weeks (311.5±7.8 vs. 412.3±9.4, P<0.01), and 12 weeks (288.1±12.1 vs. 506.1±8.9, P<0.001) after injection (Figure 1A). The level of blood glucose in the STZ-treated group was as high as that in the control group (5.31±0.78 vs. 5.21±0.68, P=0.612) at 1 week after treatment. However, this was significantly higher in the STZ-treated group than in the control group at 4 weeks (21.69±4.43 vs. 5.28±0.53, P<0.001), 9 weeks (24.50±3.43 vs. 5.10±0.86, P<0.001), and 12 weeks (26.35±4.30 vs. 5.17±0.89, P<0.001) after injection (Figure 1B).
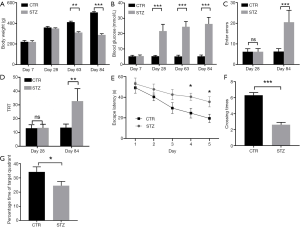
After the model was established, cognitive ability was assessed by Y-maze 4 weeks and 12 weeks after STZ treatment. At 4 weeks after treatment, the number of error times of rats from the STZ-treated group was comparable to that in the control group (6.21±1.52 vs. 6.19±1.47, P=0.971). However, at 12 weeks after treatment, the number of errors was significantly higher in the STZ-treated group than in the control group (20.52±5.81 vs. 6.27±1.43, P<0.001) (Figure 1C). The total reaction time of the Y maze of rats in the STZ-treated group was as high as that in the control group (13.28±2.68 vs. 13.04±2.52, P=0.738) 4 weeks after treatment. However, at 12 weeks after treatment, the total reaction time of the Y maze of rats in the STZ-treated group was more than that in the control group (32.76±9.03 vs. 13.46±2.62, P<0.01) (Figure 1D). These results indicate rats treated with STZ had impaired cognitive ability compared with the control group.
The Morris water maze test was then used to assess the learning ability of rats in both groups. As the training progressed, the rats of both groups showed a reduced escape latency. More importantly, on each day of the training test, STZ-treated rats exhibited much longer escape latency (24.21±4.19 vs. 42.04±9.44, P<0.05) than the control rats, suggesting an impaired learning ability of the STZ-treated rats (Figure 1E). In the spatial probe test, the STZ-treated rats had a significantly decreased number of target platform crossings compared with the control rats (2.62±1.52 vs. 6.31±1.79, P<0.001) (Figure 1F). On the other hand, the STZ-treated rats had a shorter duration time in the acquisition quadrant compared with the control rats (22.62±1.52 vs. 32.14±3.66, P<0.05) (Figure 1G). In this context, the STZ-treated rats exhibited impaired learning ability.
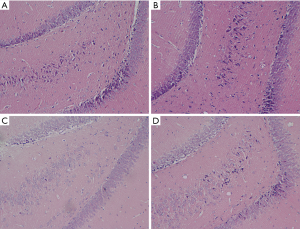
The HE staining of the hippocampi examined under light microscope revealed that hippocampal neurons in control rats were uniformly aligned, with clear nuclei staining and a transparent cytoplasm (Figure 2A,B). However, in the STZ-treated rats, some hippocampal neurons exhibited apoptotic morphology of crescent or serrated shapes (Figure 2C,D). These results indicated that STZ-treated induced loss or degeneration of hippocampal neurons.
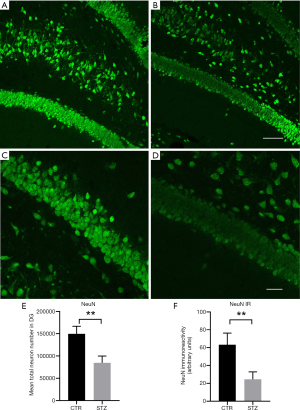
Confocal micrographs in the DG stained for NeuN of hippocampal slices between the controls (Figure 3A) and STZ-treated rats (Figure 3B). A higher magnification for NeuN staining in the controls (Figure 3C) and STZ rats (Figure 3D). By comparing the quantity of NeuN + cells (Figure 3E) and the immunoreactivity of NeuN (Figure 3F) in DG between the two groups, we found that TZ-treated induced loss or degeneration of hippocampal neurons.
STZ-induced diabetes upregulates the expression of LncRNA vof16 in the hippocampus
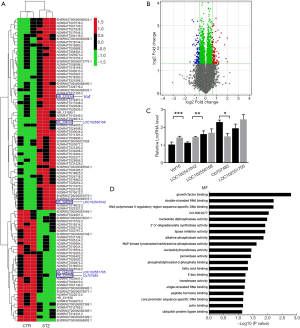
To explore the potential mechanism of reduced learning ability in STZ-induced rats, Agilent Rat lncRNA V3 was used to screen the differentiation expression of lncRNA between STZ-treated and the control rats, with a fold-change >2 identified as a differential expressed lncRNA. This showed 60 lncRNAs were upregulated in the hippocampus of STZ-treated rats, while 35 were downregulated (Figure 4A). The result also showed that five lncRNAs, LOC102547042, Vof-16, LOC102550168, Cb707485, and LOC102551705, were over 10-fold upregulated in the hippocampus of STZ-treated rats (Figure 4B). For further validation, the expression of the five differential lncRNAs was assessed with quantitative real-time PCR (qRT-PCR). This showed that the expression of Vof-16 was significantly higher in the STZ-treated group than in the control group (Figure 4C). As an ischemia related factor, vof-16 (Vof-16) has been reported to be upregulated upon hypoxic-ischemic encephalopathy injury. Additionally, Go enrichment for molecular function according to the altered lncRNA showed that the majority of differential LncRNA target genes were associated with the nucleotide diphosphatase activity, nucleotidyltransferase activity, as well as the neuron growth factor binding (Figure 4D).
LncRNA Vof-16 binds with miR-205 and functions as a ceRNA
Given that LncRNA work as ceRNA could competitively bind with miRNAs and thereby lead to the depression of miRNAs targets, we sought to identify the potential targets of LncRNA Vof-16. In brief, miR-205 was found to share the same microRNA response element with Vof-16 in silico analysis carried by both miR and an miRBase (Figure 5A). To evaluate the relationship between Vof-16 and miR-205 in vitro, Vof-16 was overexpressed in primary cultured rat hippocampal neurons and the expression of miR-205 was assessed by qRT-PCR. This showed that compared with the control group, overexpression of Vof-16 induced a decrease of miR-205, suggesting that Vof-16 may interact with miR-205 through MRE (Figure 5B).
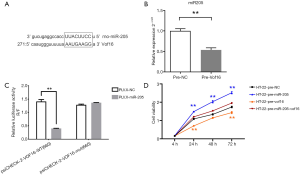
Additionally, a luciferase reporter assay was conducted in 293T cells to validate the direct binding between Vof-16 and miR-205. Firstly, the wild type (WT) and mutant (mut) Vof-16 sequence containing the predictive recognition site of miR-205 were cloned into the psiCHECK-2 vector, while miR-205 was cloned into the pLVX-Puro vector. The luciferase assay showed that transfection of pLVX-miR-205 could significantly reduce luciferase activity of Vof-16-WT, as compared with that of pLVX. However, the effect on Vof-16-mut luciferase activity of pLVX-miR-205 transfection was comparable with that of pLVX, suggesting miR-205 could efficiently bind with LncRNA Vof-16 (Figure 5C).
LncRNA Vof-16 regulates hippocampal neuron proliferation via miR-205
As diabetic-induced hippocampus neuronal degeneration is probably due to a neuron proliferation defect, we sought to assess the effects of Vof-16 and/or miR-205 on primary hippocampal neuron proliferation. The primary cultured hippocampus neurons were transfected with Vof-16, and the cell proliferation was assessed 4, 24, 48, and 72 h after transfection. CCK-8 assays revealed that the elevated expression of miR-205 significantly promoted the proliferation of hippocampus neurons at 24, 48, and 72 h after transfection, as compared with the control group. On the other hand, the overexpression of Vof-16 inhibited cell proliferation at 24, 48, and 72 h after transfection. Furthermore, the proliferation defect induced by Vof-16 elevation could be partially relieved by miR-205 co-expression at 24 and 72 h post-transfection (Figure 5D). These results indicated that Vof-16 regulates the proliferation of hippocampus neuron cells through antagonism with miR-205.
LncRNA Vof-16 plays a ceRNA role in regulating Gnb3 by binding with miR-205
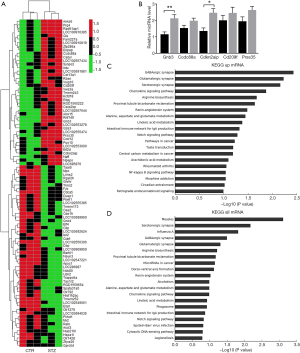
To screen the potential target of LncRNA Vof-16, the chip was used to assess the differential mRNA between control rats and STZ-treated rats and a fold-change >2 was identified as a differential expressed mRNA. As a result, 43 mRNAs were upregulated in the hippocampus of STZ-treated rats, while 52 were downregulated (Figure 6A). Among these, guanine nucleotide binding protein beta polypeptide 3 (Gnb3) was upregulated and this upregulation was confirmed by qRT-PCR (Figure 6B). Additionally, GO enrichment analyzed with the Kyoto Encyclopedia of Genes and Genomes (KEGG) showed that the majority of differential mRNA target genes were associated with the GABAergic synapse, glutamatergic synapse, as well as the neuron growth-related NF-κB signaling pathway (Figure 6C,D). These results revealed that Gnb3 is upregulated in the hippocampus of rats suffering from diabetes.
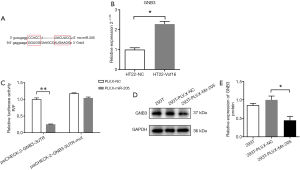
The in silico analysis showed that Gnb3 is a potential target of miR-205 as they share the same MRE (Figure 7A), and the overexpression of Vof-16 in primary rat hippocampal neurons significantly increased Gnb3 mRNA expression (Figure 7B). Therefore, a dual luciferase assay was first carried out to experimentally determine the relationship between Gnb3 and miR-205. For this purpose, Gnb3-3'-UTR containing a wild type potential binding site of miR-205 (Gnb3-3'UTR-WT) or the mutant variety (Gnb3-3'UTR-mut) was subcloned into the psiCHECK-2 vector and luciferase assay revealed that the expression of pLVX-miR-205, rather than pLVX, could significantly reduce luciferase activity of psiCHECK-2-Gnb3-3'UTR-WT. However, the luciferase activity of psiCHECK-2-Gnb3-3'UTR-mut was not dramatically changed upon pLVX-miR-205 overexpression (Figure 7C). These results indicate that miR-205 could regulate Gnb3 directly through 3'UTR sequence. To further verify the relationship between miR-205 and Gnb3, the protein level of Gnb3 was evaluated with Western Blot upon miR-205 overexpression and as expected, the overexpression of pLVX-miR-205 could significantly reduce the protein level of Gnb3 in 293T cell, as compared with pLVX (Figure 7D,E).
Discussion
Previous studies have implicated a relationship between protein-coding RNAs and non-coding RNAs and that long non-coding RNA could work as a natural sponge of endogenous microRNAs. This makes the lncRNA-miRNA-mRNA network crucial for the regulation and development of several diseases.
LncRNA Vof-16 plays vital roles in neuropathy, and its upregulation is related to learning ability impairment (16). The present study found the lncRNA Vof-16-miR-205-Gnb3 axis in STZ-induced diabetic encephalopathy in rats. Along with the upregulation of Vof-16, G-protein b3 subunit gene (Gnb3) protein level is also increased in the hippocampal neurons of STZ-induced diabetic rats. Gnb3 is one of the subunits of G-proteins which are involved in intracellular signaling transduction. Human GNB3 polymorphisms have been studied clinically, and an induced loss-of-function of GNB3, is related with hypertension, diabetes, obesity, and atherosclerosis. GNB3 C825T is the most extensively studied polymorphism, which could result in an activated G protein and thus enhanced signal transduction and a recent study revealed that GNB overexpression in mice could induce obesity and metabolic syndrome. More importantly, several studies have linked the GNB3 C825T polymorphism with the risk of diabetic neuropathy (17-19) and although its mechanism largely unknown and the GNB3 polymorphism related to diabetes seems to be ethnic specific, it is still worth noting. GNB3 C825T is also associated with the pathogenesis of Alzheimer’s disease (AD). A neuronal cell line-based study revealed that the co-expression of ADRB1 C and GNB3 T could increase MAPK activation, β-adrenergic responsiveness, and amyloidogenesis, thus AD susceptibility (20). These studies implied that a low GNB3 expression level should be tightly regulated in diabetes and related encephalopathy.
In the adult mammalian hippocampus, latent neurons wait activation in the subgranular zone (SGZ) of the dentate gyrus (DG) (21,22) and hippocampal neuron proliferation has been demonstrated to be altered in adults upon environmental or pharmacological stimuli. The reduction of the SGZ upon VPA treatment induced cognitive deficits in mice (23) and exposure to inescapable shock (IS) significantly reduced hippocampal neuron proliferation in rats (24). Chronic metabolic diseases such as type I diabetes induced by STZ injection also significantly impaired cell proliferation in the dentate gyrus resulting in cognitive decline (25). These studies support our results that Vof-16-miR-205 participates in hippocampal neuron proliferation in STZ-induced diabetic rats.
In our study, Gnb3 was tightly regulated by miR-205. Although only one study on dogs has reported the expression of miR-205 in the hippocampus, miR-205 has been intensively studied as it is either up-modulated or down-modulated in different epithelium cancers. The aberrant expression of miR-205 participates in cell proliferation and migration regulation. Moreover, the expression of miR-205 is regulated at transcriptional level and post-transcriptional level. Overexpressed lncRNAsLINC00673 has been found in hepatocellular carcinoma and LINC00152 in renal carcinoma are able to absorb miR-205 (26). In the present study, we found that lncRNA Vof-16 worked as a ceRNA to reduce miR-205 in hippocampal neurons, and Vof-16 inhibited cell proliferation via absorbing miR-205 in vitro. This reveals miR-205 promotes cell proliferation in hippocampal neurons. Previous studies have found that miR-205 promotes cell proliferation via particular targets in several types of cancer cells. In pancreatic cancer cell lines, miR-205 targeted adenomatous polyposis coli (APC) to promote cell proliferation (27), and PTEN and SMAD4 were targeted by miR-205 in ovarian cancer cells (28). Moreover, miR-205 targeted CDK2AP1 in laryngeal squamous cells to promote cell proliferation (29). In this study, we identified Gnb3 as a target of miR-205. However, previous studies have showed that GNB3 C825T carriers, the gain-of-function allele of GNB3, are more susceptible to antigen stimulation including HBV (30) or common recall antigens (31), which may induce a higher proliferation rate of lymphocytes. Therefore, it is possible that unidentified targets of miR-205 are involved in the regulation of cell proliferation in our study and more investigation of miR-205 targets in hippocampus neuron proliferation is required.
In conclusion, lncRNA Vof-16 is upregulated in hippocampal neurons in STZ-induced diabetic rats and sponging the endogenous miR-205 inhibits the proliferation of hippocampal neurons. Although GNB3 is associated with diabetes, the mechanism of Gnb3 in STZ-induced diabetic neuropathy is still to be elucidated.
Acknowledgments
Funding: The National Natural Science Foundation of China (81800715).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-21-2016
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-21-2016
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-21-2016). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (No.: 2018-37) granted by institutional ethics board of Shanghai Tongren Hospital, in compliance with the National Institutes of Health Guide for Care and Use of Laboratory Animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Melendez-Ramirez LY, Richards RJ, Cefalu WT. Complications of type 1 diabetes. Endocrinol Metab Clin North Am 2010;39:625-40. [Crossref] [PubMed]
- Nunley KA, Ryan CM, Orchard TJ, et al. White matter hyperintensities in middle-aged adults with childhood-onset type 1 diabetes. Neurology 2015;84:2062-9. [Crossref] [PubMed]
- Chen F, Ghosh A, Hu M, et al. RAGE-NF-κB-PPARγ Signaling is Involved in AGEs-Induced Upregulation of Amyloid-β Influx Transport in an In Vitro BBB Model. Neurotox Res 2018;33:284-99. [Crossref] [PubMed]
- Martin-Peña A, Rincon-Limas DE, Fernandez-Funez P. Anti-Aβ single-chain variable fragment antibodies restore memory acquisition in a Drosophila model of Alzheimer's disease. Sci Rep 2017;7:11268. [Crossref] [PubMed]
- Heinisch JJ, Brandt R. Signaling pathways and posttranslational modifications of tau in Alzheimer's disease: the humanization of yeast cells. Microb Cell 2016;3:135-46. [Crossref] [PubMed]
- Gratuze M, Planel E. Regulation of brain insulin signaling: A new function for tau. J Exp Med 2017;214:2171-3. [Crossref] [PubMed]
- Marciniak E, Leboucher A, Caron E, et al. Tau deletion promotes brain insulin resistance. J Exp Med 2017;214:2257-69. [Crossref] [PubMed]
- Awad A, Lundqvist R, Rolandsson O, et al. Lower cognitive performance among long-term type 1 diabetes survivors: A case-control study. J Diabetes Complications 2017;31:1328-31. [Crossref] [PubMed]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10:155-9. [Crossref] [PubMed]
- Hua JT, Chen S, He HH. Landscape of Noncoding RNA in Prostate Cancer. Trends Genet 2019;35:840-51. [Crossref] [PubMed]
- Li R, Fox AH. SPArking Interest in the Long Noncoding RNA World: A New Class of 5' SnoRNA-Stabilized LncRNA that Influences Alternative Splicing. Mol Cell 2016;64:435-7. [Crossref] [PubMed]
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 2016;17:272-83. [Crossref] [PubMed]
- Li B, Mao R, Liu C, et al. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci 2018;197:122-9. [Crossref] [PubMed]
- Chen J, Huang X, Wang W, et al. LncRNA CDKN2BAS predicts poor prognosis in patients with hepatocellular carcinoma and promotes metastasis via the miR-153-5p/ARHGAP18 signaling axis. Aging (Albany NY) 2018;10:3371-81. [Crossref] [PubMed]
- Wang W, Lou W, Ding B, et al. A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging (Albany NY) 2019;11:2610-27. [Crossref] [PubMed]
- Tohda M, Watanabe H. Molecular cloning and characterization of a novel sequence, vof-16, with enhanced expression in permanent ischemic rat brain. Biol Pharm Bull 2004;27:1228-35. [Crossref] [PubMed]
- Zychma MJ, Zukowska-Szczechowska E, Ossowska-Szymkowicz I, et al. G-Protein beta(3) subunit C825T variant, nephropathy and hypertension in patients with type 2 (Non-insulin-dependent) diabetes mellitus. Am J Nephrol 2000;20:305-10. [Crossref] [PubMed]
- Beige J, Ringel J, Distler A, et al. G-protein beta(3)-subunit C825T genotype and nephropathy in diabetes mellitus. Nephrol Dial Transplant 2000;15:1384-7. [Crossref] [PubMed]
- Blüthner M, Schmidt S, Siffert W, et al. Increased frequency of G-protein beta 3-subunit 825 T allele in dialyzed patients with type 2 diabetes. Kidney Int 1999;55:1247-50. [Crossref] [PubMed]
- Bullido MJ, Ramos MC, Ruiz-Gómez A, et al. Polymorphism in genes involved in adrenergic signaling associated with Alzheimer's. Neurobiol Aging 2004;25:853-9. [Crossref] [PubMed]
- van Praag H, Schinder AF, Christie BR, et al. Functional neurogenesis in the adult hippocampus. Nature 2002;415:1030-4. [Crossref] [PubMed]
- Seri B, García-Verdugo JM, McEwen BS, et al. Astrocytes give rise to new neurons in the adult mammalian hippocampus. J Neurosci 2001;21:7153-60. [Crossref] [PubMed]
- Umka J, Mustafa S, ElBeltagy M, et al. Valproic acid reduces spatial working memory and cell proliferation in the hippocampus. Neuroscience 2010;166:15-22. [Crossref] [PubMed]
- Malberg JE, Duman RS. Cell proliferation in adult hippocampus is decreased by inescapable stress: reversal by fluoxetine treatment. Neuropsychopharmacology 2003;28:1562-71. [Crossref] [PubMed]
- Jackson-Guilford J, Leander JD, Nisenbaum LK. The effect of streptozotocin-induced diabetes on cell proliferation in the rat dentate gyrus. Neurosci Lett 2000;293:91-4. [Crossref] [PubMed]
- Ferrari E, Gandellini P. Unveiling the ups and downs of miR-205 in physiology and cancer: transcriptional and post-transcriptional mechanisms. Cell Death Dis 2020;11:980. [Crossref] [PubMed]
- Qin RF, Zhang J, Huo HR, et al. MiR-205 mediated APC regulation contributes to pancreatic cancer cell proliferation. World J Gastroenterol 2019;25:3775-86. [Crossref] [PubMed]
- Chu P, Liang A, Jiang A, et al. miR-205 regulates the proliferation and invasion of ovarian cancer cells via suppressing PTEN/SMAD4 expression. Oncol Lett 2018;15:7571-8. [Crossref] [PubMed]
- Zhong G, Xiong X. miR-205 promotes proliferation and invasion of laryngeal squamous cell carcinoma by suppressing CDK2AP1 expression. Biol Res 2015;48:60. [Crossref] [PubMed]
- Lindemann M, Barsegian V, Siffert W, et al. Role of G protein beta3 subunit C825T and HLA class II polymorphisms in the immune response after HBV vaccination. Virology 2002;297:245-52. [Crossref] [PubMed]
- Lindemann M, Virchow S, Ramann F, et al. The G protein beta3 subunit 825T allele is a genetic marker for enhanced T cell response. FEBS Lett 2001;495:82-6. [Crossref] [PubMed]
(English Language Editor: B. Draper)


