
Identification of genes predicting unfavorable prognosis in hepatitis B virus-associated hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is one of the most common primary liver cancers, accounting for the third highest number of cancer-associated deaths worldwide (1,2). Multiple factors including hepatitis, diabetes, smoking, and alcohol consumption are known as risk factors of HCC (3,4). Among them, hepatitis B virus (HBV) infection serves as the leading cause contributing to the development and progression of HCC, which has been found to be related to 66% of cases (5). Although there have been great advances in HCC diagnosis and surgical techniques, patients with HBV-associated HCC have poor clinical prognosis due to virus induced-genetic alterations and irreversible hepatic damage and cirrhosis (6,7). Therefore, further identification of genomic alterations of HBV-associated HCC is essential to provide potential targets for early diagnosis, as well as to develop novel therapeutic strategies.
Over the past decades, gene profiling and signatures, which can quickly detect differentially expressed genes (DEGs), have greatly accelerated cancer research. Several studies have analyzed the prognostic effects of array-based genes from HCC tumors. With genome-wide expression profiling, Cai et al. (8) developed a signature consisting of 11 genes that could effectively predict the overall survival (OS) of postoperative HCC patients. In addition, a 7-miRNAs-based signature was found to be significantly associated with recurrence-free survival in HCC (9). However, few investigations have identified gene signatures that predict poor prognosis for HBV-associated HCC. Thus, with public massive data and integrated bioinformatics methods, identifying important genes to predict prognosis in HBV-associated HCC is necessary and of great clinical significance (10).
With increasing use of high-throughput techniques, Gene Expression Omnibus (GEO) provides a public gene expression platform that contains millions of datasets and samples. It facilitates gene analysis including biomarker discovery, disease classification and phenotype comparisons. In the present study, we first reviewed HCC datasets in Gene Expression Omnibus (GEO), and 3 eligible datasets including patients with HBV-associated HCC were selected. Through overlapping analysis, a gene set with DEGs was identified. Of these DEGs, 14 were identified as hub DEGs in the protein-protein interaction (PPI) network. Different from previous studies identifying key biomarkers lacking of validation (11), we conducted validation analysis using GSE14520 clinical data which included HBV-associated tumor samples with complete prognostic information. The survival analysis of hub DEGs demonstrated that only high expression of TOP2A was significantly associated with both poor OS and disease-free survival (DFS). Therefore, TOP2A may serve as a biomarker for prognosis assessment and as a therapeutic target for HBV-associated HCC.
We present the following article in accordance with the REMARK reporting checklist (available at https://dx.doi.org/10.21037/atm-21-2085).
Methods
Gene datasets
The NCBI-GEO databases were searched to identify datasets that determined the DEGs in HCC. Eligible gene expression profiles of GSE 121248, GSE 62232, and GSE 55092 containing HBV-associated HCC and adjacent normal liver tissues were obtained. Microarray data of GSE121248, GSE62232, and GSE55092 were all based on GPL570 Platforms [(HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array] which included 70 HBV-associated HCC tissues and 37 normal tissues, 10 HBV-associated HCC tissues and 10 normal tissues, and 39 HBV-associated HCC tissues and 81 normal tissues, respectively. All procedures were in accordance with the ethical standards of institutional and national committee on human experimentation and with the Helsinki Declaration (as revised in 2013).
Identification of DEGs
The genome expression profile was compared in the R platform using the limma package (12). Genes with |logFC| >2 and adjusted P value <0.05 between HBV-associated HCC and normal liver tissues were identified as DEGs. Volcano plots were constructed with the gplots package and overlapping genes were obtained via the online Venn diagram analysis.
Analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways
GO is a community-based bioinformatics resource that supplies information regarding unique gene properties (13). KEGG is an integrated database resource for the biological interpretation of genomes, compounds, enzymes, diseases, drugs, and biological pathways. The online bioinformatics tool DAVID (14) was used to analyze GO enrichment and KEGG pathways (P<0.05).
Construction of the PPI network
For DEGs in the 3 cohorts, PPI networks for these genes were constructed with the online tools STRING and Cytoscape software (15). Moreover, the MCODE app in Cytoscape was used to assess the node distribution, path length distribution, and average clustering distribution. The top nodes with the highest degree of connectivity were identified as hub genes.
Survival analysis and identification of hub genes
In order to validate the DEGs that were associated with the prognosis of HCC patients, hub genes in HBV-associated HCC were further assessed using GSE14520 clinical data. The OS and DFS of each hub gene were determined by Kaplan-Meier analysis. Finally, the correlation between clinicopathological parameters and significant hub genes was evaluated.
Statistical analysis
The test used to compare the expression between two groups was assessed by independent sample t-test. The FDR in DEG screening and GSEA were performed according to the Benjamini-Hochberg procedure. Volcano plots were constructed with the gplots package and overlapping genes were obtained via the online Venn diagram analysis. All statistical analyses were performed using SPSS, version 24.0 software with P<0.05 defined as statistical significance.
Results
Identification of DEGs in HBV-associated HCC
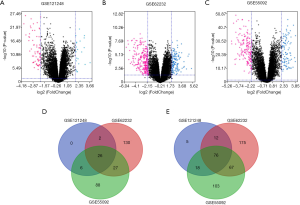
A total of 119 HBV-associated HCC tissues and 128 adjacent normal tissues from 3 datasets were included in our study. The whole-genome expression profile was compared in the R platform by using the limma package. A total of 145, 515, and 411 DEGs from GSE 121248, GSE 62232, and GSE 55092 were detected, respectively. The volcano plots of the DEGs are shown in Figure 1A,B,C. A Venn diagram was then used to obtain the overlapping genes within DEGs (Figure 1D,E). Finally, 102 overlapping genes were identified in HBV-associated HCC, including 26 up-regulated genes and 76 down-regulated genes (Table 1).

Full table
Functional evaluation of DEGs
The online bioinformatics tool DAVID software was then used to investigate 102 DEGs in terms of GO enrichment and KEGG pathways. The GO analysis demonstrated that up-regulated DEGs were significantly enriched in regulation of cell division, M phase, nuclear division, and mitosis for biological processes (BP), while down-regulated DEGs were enriched in oxidation reduction, secondary metabolic process, innate immune response, vitamin metabolic process, and immune response. For molecular function (MF), centrosome, microtubule organizing center, cytosol, and protein serine played the major roles in up-regulated DEGs, while electron carrier activity, iron ion, heme, tetrapyrrole, sugar, and carbohydrate binding were involved in down-regulated DEGs. In addition, spindle, cytoskeleton, and intracellular non-membrane-bounded organelle were the most significant cell components (CC) in up-regulated DEGs. Down-regulated DEGs were enriched in extracellular region, cell fraction, intrinsic and integral to plasma membrane, and endomembrane system (Figure 2A,B and Tables S1,S2).
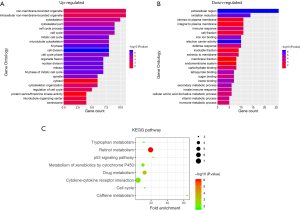
In line with GO term enrichment results, KEGG analysis showed that DEGs were mainly responsible for metabolism-related biological processes including retinol, drug, caffeine, cytochrome P450, and tryptophan metabolism. P53 pathway and cytokine-cytokine receptor interaction were also involved in the development of HBV-associated HCC by DEGs (Figure 2C and Table 2).

Full table
Construction of the PPI network
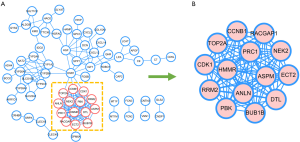
To analyze the PPI, the selected 102 DEGs were covered. With the Cytoscape software, a PPI network was constructed with 70 nodes and 175 edges (Figure 3A). The top-ranked linker nodes included TOP2A, HMMR, DTL, CCNB1, NEK2, PBK, RACGAP1, PRC1, CDK1, RRM2, ECT2, BUB1B, ANLN, and ASPM via the MCODE app (Figure 3B). A simplified PPI network with the 14 up-regulated genes was generated. The selected hub genes in the cluster may have prognostic value for HBV-associated HCC.
Validation of hub genes
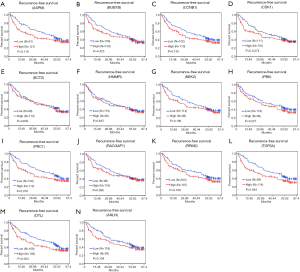
To confirm the effect of genes on survival, the 14 hub genes were further evaluated using GSE14520 clinical data. A total of 212 HBV-associated tumor samples and 210 adjacent normal samples with complete prognostic information were included. High expression of ASPM, BUB1B, CDK1, NEK2, PBK, PRC1, RACGAP1, RPM2, TOP2A, ANLN, and DTL were significantly associated with an increased risk of death (Figure S1). However, only patients with overexpression of TOP2A showed unfavorable recurrence-free survival (Figure 4). Further analysis based on age, gender, AFP level, tumor size, grade, and numbers in the GSE14520 cohort indicated that high expression of TOP2A was associated with an increased level of AFP (Figure 5 and Table 3).
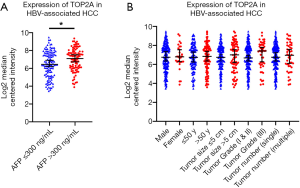
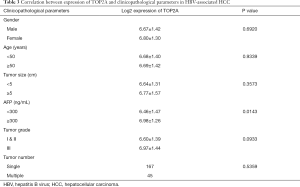
Full table
Discussion
According to the estimates of recent study, about 350 million people worldwide are infected with HBV. HBV infection, the leading etiology of HCC, accounts for 66% cases of HCC incidence globally (16). HBV infection is mainly transmitted vertically in endemic areas, especially in developing countries. Accordingly, the average age of HBV carriers who develop HCC is younger than that of other etiologies. Besides, liver inflammation caused by immune responses during HBV infection leads to liver fibrosis and cirrhosis in majority of patients, which promotes the development of HCC. In addition, integration of HBV DNA into the host genome induces both genetic instability and mutagenesis of various cancer-related genes, which is pathologically unique compared with other types of HCC (17,18). Since the key genes of HBV-associated HCC are different from other types of HCC, we conducted this study to explore DEGs for their novel prognostic value and clinical significance in HBV-associated HCC.
In the present study, we identified 26 up-regulated genes and 76 down-regulated genes which were involved in cell division, metabolism-related biological processes, the p53 pathway, and the cell cycle. Additionally, 14 hub DEGs were further validated and TOP2A was found to be associated with unfavorable OS and recurrence-free survival in HBV-associated cohorts. TOP2A, also known as DNA topoisomerase II alpha, is highly expressed at G2/M phase and a key regulator of DNA decatenation during mitosis (19,20). As a marker of proliferation in malignant cells, TOP2A has been found correlated with poor prognosis in various types of cancers such as prostate cancer, breast cancer, and gallbladder carcinoma (21-23). Our results demonstrated higher expression of TOP2A in HBV-associated HCC tissues and worse survival outcomes induced by TOP2A. The results were consistent with previous studies in HCC (24,25). Recent study by Kwan et al. (26) showed that TOP2A was a downstream target of TRRAP and KAT regulating HCC cell growth. Depletion of TOP2A caused reduced colony formation, induction of senescence and G2/M arrest of HCC, which confirmed the cancer promoting role of TOP2A. Panvichian et al. further reported that TOP2A in HCC was related to the presence of hepatitis B surface antigen in serum (27). This may explain why TOP2A was uniquely identified from hub genes in HBV-associated HCC. In addition, an elevated level of AFP was also found in patients with high TOP2A expression, which is in line with other malignant characteristics including microvascular invasion and chemotherapy resistance reported by other studies (28). Thus, high expression of TOP2A may serve as one of the genomic hallmarks for prognosis and as a therapeutic target for HBV-associated HCC.
Recently, multiple studies have identified crucial genes for HCC. DEGs including CCNB1, CDC20, PRC1, NDC80, and FOXM1, among others, were found to be up-regulated in tumor tissues and predicted poor prognosis (29,30). The difference between our study and previous studies was that we focused on a genetically different type of HCC. Our current study selected gene profiles from HBV-associated tissues, while other studies included HCC samples which originated from diverse etiologies including hepatitis infection, autoimmune disease, and nonalcoholic steatohepatitis, which resulted in different gene signatures (31). In addition, another advantage in our study was that hub genes identified by the PPI network were further validated in the biggest online HBV-associated cohort based on GSE14520. TOP2A, with significant prognostic value, was revealed by survival analysis, which is more reliable compared with previous studies.
Certain underlying limitations need to be considered when interpreting our results. Firstly, our study only enrolled 3 gene expression profiles from GEO datasets comprising 119 HCC tissues and 128 adjacent normal tissues. The 3 GEO datasets lack expressions of microRNA, lncRNA and circRNA as well. The limited availability of HBV-associated HCC datasets impeded us from accurately screening DEGs. Also, due to the absence of HBV infection status in TCGA datasets, our current study failed to validate hub genes in TCGA cohorts. In addition, clinicopathological parameters from GSE14520 including age, gender, tumor size, grade, and number were not sufficient. More tumor characteristics are required to evaluate the prognostic value of key genes. Besides, the basic experimental data validating the role of TOP2A is lacking. Further experiments both in vivo and in vitro are essential to confirm the prognostic value of TOP2A in HBV-associated HCC.
Conclusions
Despite underlying limitations, our present study identified 102 DEGs in HBV-associated HCC. GO and KEGG analysis demonstrated increased division and proliferation of HBV-associated cancer cells induced by these DEGs. Using the PPI network and further investigations, we found that overexpression of TOP2A was significantly associated with poor prognosis and increased AFP level. TOP2A might serve as a key gene for prognosis and as a therapeutic target for HBV-associated HCC.
Acknowledgments
Funding: This study was supported by National Key Research on Precision Medicine of China (2017YFC0908102, 2018ZX10723204) and National Natural Science Foundation of China (81902379).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-2085
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-2085). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures were in accordance with the ethical standards of institutional and national committee on human experimentation and with the Helsinki Declaration (revised in 2013). There was no interaction with patients directly, as we acquired data from online public datasets.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301-14. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Wang M, Xi D, Ning Q. Virus-induced hepatocellular carcinoma with special emphasis on HBV. Hepatol Int 2017;11:171-80. [Crossref] [PubMed]
- Fujiwara N, Friedman SL, Goossens N, et al. Risk factors and prevention of hepatocellular carcinoma in the era of precision medicine. J Hepatol 2018;68:526-49. [Crossref] [PubMed]
- Stanaway JD, Flaxman AD, Naghavi M, et al. The global burden of viral hepatitis from 1990 to 2013: findings from the Global Burden of Disease Study 2013. Lancet 2016;388:1081-8. [Crossref] [PubMed]
- Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, et al. Hepatocellular carcinoma: From diagnosis to treatment. Surg Oncol 2016;25:74-85. [Crossref] [PubMed]
- Genco C, Cabibbo G, Maida M, et al. Treatment of hepatocellular carcinoma: present and future. Expert Rev Anticancer Ther 2013;13:469-79. [Crossref] [PubMed]
- Cai J, Li B, Zhu Y, et al. Prognostic Biomarker Identification Through Integrating the Gene Signatures of Hepatocellular Carcinoma Properties. EBioMedicine 2017;19:18-30. [Crossref] [PubMed]
- Wang G, Dong F, Xu Z, et al. MicroRNA profile in HBV-induced infection and hepatocellular carcinoma. BMC Cancer 2017;17:805. [Crossref] [PubMed]
- Roessler S, Long EL, Budhu A, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology 2012;142:957-966.e12. [Crossref] [PubMed]
- Liao X, Yu T, Yang C, et al. Comprehensive investigation of key biomarkers and pathways in hepatitis B virus-related hepatocellular carcinoma. J Cancer 2019;10:5689-704. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47 [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Kulik L, El-Serag HB. Epidemiology and Management of Hepatocellular Carcinoma. Gastroenterology 2019;156:477-491.e1. [Crossref] [PubMed]
- Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol 2019;16:589-604. [Crossref] [PubMed]
- Schweitzer A, Horn J, Mikolajczyk RT, et al. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 2015;386:1546-55. [Crossref] [PubMed]
- D Arcy N. Gabrielli B. Topoisomerase II Inhibitors and Poisons, and the Influence of Cell Cycle Checkpoints. Curr Med Chem 2017;24:1504-19. [PubMed]
- Delgado JL, Hsieh CM, Chan NL, et al. Topoisomerases as anticancer targets. Biochem J 2018;475:373-98. [Crossref] [PubMed]
- Labbé DP, Sweeney CJ, Brown M, et al. TOP2A and EZH2 Provide Early Detection of an Aggressive Prostate Cancer Subgroup. Clin Cancer Res 2017;23:7072-83. [Crossref] [PubMed]
- Washiro M, Ohtsuka M, Kimura F, et al. Upregulation of topoisomerase IIalpha expression in advanced gallbladder carcinoma: a potential chemotherapeutic target. J Cancer Res Clin Oncol 2008;134:793-801. [Crossref] [PubMed]
- Romero A, Martín M, Cheang MC, et al. Assessment of Topoisomerase II α status in breast cancer by quantitative PCR, gene expression microarrays, immunohistochemistry, and fluorescence in situ hybridization. Am J Pathol 2011;178:1453-60. [Crossref] [PubMed]
- Zhang L, Huang Y, Ling J, et al. Screening and function analysis of hub genes and pathways in hepatocellular carcinoma via bioinformatics approaches. Cancer Biomark 2018;22:511-21. [Crossref] [PubMed]
- Wu M, Liu Z, Zhang A, et al. Identification of key genes and pathways in hepatocellular carcinoma: A preliminary bioinformatics analysis. Medicine (Baltimore) 2019;98:e14287 [Crossref] [PubMed]
- Kwan SY, Sheel A, Song CQ, et al. Depletion of TRRAP Induces p53-Independent Senescence in Liver Cancer by Down-Regulating Mitotic Genes. Hepatology 2020;71:275-90. [Crossref] [PubMed]
- Panvichian R, Tantiwetrueangdet A, Angkathunyakul N, et al. TOP2A amplification and overexpression in hepatocellular carcinoma tissues. Biomed Res Int 2015;2015:381602 [Crossref] [PubMed]
- Wong N, Yeo W, Wong WL, et al. TOP2A overexpression in hepatocellular carcinoma correlates with early age onset, shorter patients survival and chemoresistance. Int J Cancer 2009;124:644-52. [Crossref] [PubMed]
- Shen S, Kong J, Qiu Y, et al. Identification of core genes and outcomes in hepatocellular carcinoma by bioinformatics analysis. J Cell Biochem 2019;120:10069-81. [Crossref] [PubMed]
- Wang J, Tian Y, Chen H, et al. Key signaling pathways, genes and transcription factors associated with hepatocellular carcinoma. Mol Med Rep 2018;17:8153-60. [Crossref] [PubMed]
- Zhou L, Du Y, Kong L, et al. Identification of molecular target genes and key pathways in hepatocellular carcinoma by bioinformatics analysis. Onco Targets Ther 2018;11:1861-9. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


