
Tumor protein D52 promotes breast cancer proliferation and migration via the long non-coding RNA NEAT1/microRNA-218-5p axis
Introduction
Breast cancer is an aggressive disease with high global rates of morbidity (11.7%) and mortality (6.9%) among women (1). Despite the improvement of diagnostic and therapeutic strategies for breast cancer in recent years, the prognostic and survival outcomes of patients with breast cancer are still not satisfactory. The abnormal proliferation and metastasis of tumor cells is the main cause of cancer-associated mortality in patients with cancer (2). Therefore, efforts to improve breast cancer treatment outcomes are necessary.
Studies have shown that long non-coding RNAs (lncRNAs) are important in the progression of various cancers through their post-transcriptional modulation and epigenetic modification, and the occurrence of human malignancies frequently accompanies lncRNA dysregulation (3). The upregulation of LINC00673 has been reported to be related to a poor prognosis in patients with breast cancer (4). Nuclear enriched abundant transcript 1 (NEAT1) is a lncRNA that is localized to nuclear paraspeckles. Previous studies have shown that NEAT1 can inhibit microRNA (miRNA/miR)-133b to promote breast cancer cell migration and invasion (5), and that it also participates in breast cancer progression by regulating miR-448 (6).
Tumor protein D52 (TPD52), which is upregulated in prostate and breast cancer, plays an important role in cell proliferation, migration, and death. Specifically, a previous study on neuroblastoma showed that TPD52 exerted a protective effect against apoptosis and arrested cell proliferation by increasing the expression level of p27Kip1 and activating Akt and ERK1/2, thereby promoting cell differentiation (7). In prostate cancer, miR-224 can target TPD52 to inhibit cell migration and invasion (8). TPD52 has been widely reported to be upregulated in various cancers, as have its regulatory functions in different signaling pathways (9,10). For instance, one study showed that TPD52 promotes the proliferation of breast cancer cells by promoting GSK3β phosphorylation (11). However, in another study, the overexpression of TPD52 in renal cell carcinoma cells resulted in suppression of the PI3K/AKT pathway (12). These studies indicate that TPD52 regulates the PI3K/AKT pathway, which participates in tumor cell proliferation, migration, and survival, and this may be a mechanism underlying the effects of TPD52 in cancer .
lncRNAs can act as a competitive endogenous RNAs (ceRNA) by affecting miRNAs and inhibiting their expression, resulting in the upregulation of their target genes (13). The current study aimed to investigate the direct and predicted interactions between NEAT1, miR-218-5p, and TPD52 in breast cancer. We hypothesized that TPD52 and NEAT1 are ceRNAs that compete to bind with miR-218-5p. The effects of TPD52 knockdown on breast cancer cells was assessed applying MDA-MB-231 and BT474 cell lines in vitro and vivo. Bioinformatics analysis was performed to predict the binding of TPD52 and NEAT1 to miR-218-5p. Furthermore, the expression of NEAT1 and miR-218-5p in breast cancer cells was evaluated, and the hypothesis that NEAT1 acts as a modulator of TPD52 expression by sponging miR-218-5p in breast cancer was examined. We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-2668).
Methods
Cell culture
The breast cancer cell lines BT474, MCF-7, MDA-MB-453, and MDA-MB-231 were obtained from the Shanghai Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 Medium (Thermo Fisher Scientific) supplemented with 100 U/mL penicillin/streptomycin and 10% fetal bovine serum (FBS; Thermo Fisher Scientific). The normal breast epithelial cell line MCF10A was also obtained from the same institution. The MCF10A cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM)/F12 (Thermo Fisher Scientific), which also supplemented with 10% FBS, 20 ng/mL EGFR (PeproTech Inc.), 0.5 mg/mL hydrocortisone (Sigma), 1 mg/mL cholera toxin (Enzo Life Sciences, Inc.), 10 µg/mL insulin (Sigma; Merck KGaA), and 2 mM L-glutamine (Sigma; Merck KGaA). All cells were cultured in a humidified incubator.
Cell transfection
We purchased pcDNA3.1-NEAT1 and pcDNA3.1-TPD52 plasmids and the pcDNA3.1-vector from Guangzhou RiboBio Co., Ltd . Small interfering RNA (siRNA) targeting NEAT1 (si-NEAT1-1, 5'-CTCCTTTTGGACTTTTCTCTAGG-3' and si-NEAT1-2, 5'-GTGATTGCATTGCAGATTACTAG-3'); TPD52 (si-TPD52-1, 5'-GAGTGAACAAAAGCTATCTCTAC-3' and si-TPD52-2, 5'-TTGAAGAAAAGGTCGAAAACTTA-3'); miR-218-5p mimics (5'-UUGUGCUUGAUCUAACCAUGU-3'); miR-218-5p inhibitor (5'-ACAUGGUUAGAUCAAGCACAA-3'); and their respective controls (mimics control, 5'-UUACUCGACACGUGUCAAGUUU-3'; and inhibitor control, 5'-CAGUACUUUUGUGUAGUACAA-3'), with unrelated sequences, were also synthesized by Guangzhou RiboBio Co., Ltd. Lipofectamine® 3000 (Thermo Fisher Scientific) was used to transiently transfect overexpression plasmids (1,000 ng/6 cm dish) and siRNAs (30 nM) into cells, which were subsequently cultured in a humidified incubator for 36 hours. Experiments were performed 36 hours after transfection. Si-NEAT1-2 and si-TPD52-2 were selected, as they have higher interference efficiency than si-NEAT1-1 and si-TPD52-1, respectively.
Analysis of data from The Cancer Genome Atlas (TCGA) and binding sites
TCGA contains data of breast cancer cases, which can be downloaded from the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). We performed a database search of RNASeq V2 data using the term “breast cancer”. For each case, the expression values of miR-218-5p, TPD52, and NEAT1 were collected. Pearson’s correlation test was employed to analyze the correlation between the expression of miR-218-5p and the expressions of TPD52 and NEAT1. The StarBase database (http://starbase.sysu.edu.cn/) was employed to search for miR-218-5p binding sites on TPD52 and NEAT1.
Quantitative reverse transcription (RT)-polymerase chain reaction
Total cellular RNA was collected with TRIzol® (Thermo Fisher Scientific). Complementary DNA was synthesized using a PrimeScript™ RT kit (Takara). After that, quantitative polymerase chain reaction (qPCR) of each sample was performed using TB Green® Fast qPCR Mix (Takara), with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) serving as the endogenous control. A 2-step cycling condition was selected for RT-qPCR (14). The primers used are shown in Table S1.
Western blot analysis
After being washed with cold phosphate-buffered saline (PBS), cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (Biotechnology). The collected lysates were centrifuged at 12,000 ×g at 4 °C for 10 minutes, and then the total protein was quantified using a commercial bicinchoninic acid (BCA) kit (Biotechnology). Then, 30 µg of protein was loaded into each lane, and the protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 8–12%), and transferred to a polyvinylidene difluoride (PVDF) membrane (EMD Millipore). Then, the membrane was blocked with skimmed milk and incubated overnight with the following antibodies: anti-TPD52 (1:10,000; Abcam), anti-Ki-67 (1:5,000; Abcam), anti-MCM2 (1:1,000; Abcam), anti-proliferating cell nuclear antigen (1:1,000; Cell Signaling Technology), and anti-GAPDH (1:10,000; Proteintech Group). After that, the membrane was washed 3 times with 1% tris-buffered saline (TBS) and the following secondary antibodies were added: horse-radish peroxidase (HRP) conjugated Goat Anti-Mouse IgG (1:5,000; Wuhan Boster Biological Technology) and HRP-conjugated Goat Anti-Rabbit IgG (1:5,000; Wuhan Boster Biological Technology). The protein bands were detected with a BeyoECL Plus kit (Biotechnology) and quantified by ImageJ.
Colony formation assay
Transfected cells were subjected to 2 weeks of culture in 6-well plates. The formed colonies were then fixed with 4% paraformaldehyde and stained with 0.1% crystal violet (Beijing Solarbio Science and Technology). Images were captured, and the number of colonies was counted under an optical microscope.
Cell migration assay
To evaluate the ability of cells to migrate in vitro, Transwell assays were performed as previously described (15). A total of 2×104 cells in RPMI-1640 Medium (Thermo Fisher Scientific) were seeded in the top chamber (8-µm pore; Corning, Inc.), and 600 µL culture medium, supplemented with 10% FBS as a chemoattractant, was added to the lower chamber. After incubation at 37 °C for 48 hours, the cells remaining in the top chamber were removed, while the migrated cells were fixed with 4% paraformaldehyde for 25 minutes and stained with crystal violet for 5 minutes. Images were captured of each well, and the cells in 6 random fields of view were counted using an inverted light microscope (Olympus Corporation).
Wound healing assay
BT474 or MDA-MB-231 cells were cultured in 6-well plates. After starvation in serum-free RPMI-1640 (Thermo Fisher Scientific) medium for 24 hours, a straight scratch was made at the bottom of the plate. Then, after gentle rinsing, the cells were cultured in RPMI-1640 medium for 24 hours. An inverted optical microscope was used to observe the cells, and cell migration was calculated at 0 and 24 hours. Scratch-healing (%) = (initial-final) scratch area/initial scratch area ×100.
5'-Ethynyl-2'-deoxyuridine assay
Cell proliferation was examined using a 5'-ethynyl-2'-deoxyuridine (EdU) assay. Cells were seeded in 24-well culture plates and 10 µM EdU was added to each well. After 2 hours of incubation, the cells were fixed with 4% paraformaldehyde. The incorporated EdU was detected using a kFluor488-EdU kit (Nanjing KeyGen Biotech), with incubation for 30 minutes. After staining with 4'6-diamidino-2-phenylindole (DAPI), the cells were observed under an Olympus microscope.
Dual-luciferase reporter assay
Sequences of the respective 3'-untranslated regions (UTRs) of NEAT1 and TPD52 containing miR-218-5p were subcloned into the psiCHECK2 dual-luciferase vector (Promega Corporation). Lipofectamine® 3000 (Thermo Fisher Scientific) was used to co-transfect MDA-MB-231 cells with luciferase reporter plasmid with miR-218-5p mimic or negative control (NC). The cells were then cultured in a humidified incubator with 5% CO2 at 37 °C for 36 hours, after which experiments were performed. Luciferase signals were measured using the Dual-Glo® Luciferase Assay System (Promega Corporation) according to the manufacturer’s instructions. Cells were lysed with Passive Lysis Buffer for 15 minutes, and then 20 µL cell lysate was added to a luminometer tube containing 100 µL luciferase assay reagent (LAR) II and firefly luciferase luminescence was measured. After the addition of 100 µL Stop & Glo® Reagent, Renilla luciferase luminescence was measured. A luminescence microplate reader (Tecan Group, Inc.) was used to measure the expressed luciferase activities. Firefly luciferase activity was normalized to Renilla luciferase activity and expressed as relative luciferase units to reflect the promoter activity. The binding ratio was calculated as follows: relative luciferase activity = firefly luciferase reading/Renilla luciferase reading.
In vivo tumor formation assay
All procedures performed using animals were approved by the Animal Experiments Committee of the Southern Medical University (approval Nos. L2018015 and 201690525). All applicable international, national and/or institutional guidelines for the care and use of animals were followed. Six 4- to 6-week-old nude mice were obtained from Southern Medical University and were housed under a 12-hour day/night cycle under specific pathogen-free conditions (3 companions per cage), and given standard food and water ad libitum. Short hairpin RNA (shRNA)-mediated silencing of TPD52 was performed by lentiviral infection of MDA-MB-231 cells using a pLKO-shRNA expression vector. Sh-TPD52 was used to silence TPD52, and sh-NC was used as the control. ShRNA-TPD52 lentivirus and NC lentivirus were purchased from Shanghai Genechem Co. Ltd. Lipofectamine® 3000 (Thermo Fisher Scientific) was used for transfection, and then the cells were cultured in a humidified incubator under the above conditions. The transfection efficiency was detected 48 hours after transfection. The mice were divided into 2 groups (n=3), and a disposable syringe was used to subcutaneously inject 100 µL of suspension containing 1×106 cells on the right flank of the mice (16,17). Tumor growth was observed, and the tumor volume was measured every 4 days. After 30 days, the mice were euthanized under anesthesia with 1% sodium pentobarbital (50 mg/kg), delivered via intraperitoneal injection, followed by cervical dislocation. After extraction of the tumors, images were captured and measurements were taken with a vernier caliper. The tumor volume was calculated according to the following formula: longest diameter × 2(shortest diameter) × 0.5.
Immunohistochemical staining
The xenograft tumors were fixed in 2% paraformaldehyde for 24 hours. The paraffin-embedded tissue was cut into 5-µm sections. The sections were deparaffinized, rehydrated, and then subjected to high pressure for antigen retrieval (pressure, 15PSI; temperature, 121 °C) in EDTA antigen retrieval buffer (1 mM EDTA; pH 8.0). Endogenous peroxidase was inactivated with 3% hydrogen peroxide, and 1% bovine serum albumin (Sigma) was used to block non-specific binding for 1 hour. Anti-Ki-67 (1:200; Abcam), anti-MCM2 (1:200; Abcam), and anti-PCNA (1:1,000; Cell Signaling Technology) antibodies were used to detect the expression levels of the corresponding proteins in the mouse xenograft tissues. These antibodies were the same as those used for western blotting and have been described previously. Tissue samples were incubated in a primary antibody solution overnight at 4 °C. The secondary antibody Biotin Conjugated AffiniPure Donkey Anti-Rabbit IgG (1:200; Wuhan Boster Biological Technology) was incubated for 1 hour . Finally, the tissue slices were visualized with a Nikon ECLIPSE Ti microscope system (Nikon Corporation), and the images were processed using Nikon software (NIS-Elements v.AR4.10, Nikon Corporation). IHC Profiler (an ImageJ plugin) was used to perform automatic quantitative analysis in line with the manufacturer’s instructions.
Statistical analysis
Complete randomization was used for grouping. Random numbers were generated using SAS 9.4 software (SAS Institute, Inc., Cary, NC) and then grouped using a random number generator. IBM SPSS Version 20.0 software (IBM Corp., Armonk, NY) was used for all data analysis. Results were presented as means ± standard deviations. Comparisons between multiple time points were assessed by 2-way analysis of variance. Comparisons between 2 groups at a single time point were assessed using the independent Student’s t-test. A 2-tailed P value <0.05 was considered to be statistically significant.
Results
TPD52 is upregulated in breast cancer cell lines
Using publicly available breast cancer data from the TCGA database, TPD52 and miR-218-5p were found to be negatively correlated in 744 patients with breast cancer (Figure 1A). Also, NEAT1 and miR-218-5p were negatively correlated in 422 patients with breast cancer (Figure 1B). Using the TargetScan online database, we identified the potential binding sites between TPD52 and miR-218-5p (chromosome 8, 80949405-80949411), and NEAT1 and miR-218-5p (chromosome 11, 65203519-65203539) (Figure 1C,D). We hypothesized that TPD52 and NEAT1 are ceRNAs that compete to bind with miR-218-5p in the nucleus; miR-218-5p may bind to the specific miRNA response elements of NEAT and TPD52, with binding to the latter inhibiting the translation of TPD52 or degrading TPD52, thereby reducing the TPD52 content. The lncRNA NEAT1/miR-218-5p/TPD52 axis may constitute a ceRNA network (Figure 1E). The expression levels of TPD52 were measured in 4 human breast cancer cell lines and 1 normal breast cell line. As revealed by the RT-qPCR results in Figure 1F, compared with that in MCF10A cells, the expression of TPD52 was significantly elevated in breast cancer cells, especially MDA-MB-231 cells. TPD52 protein expression was also higher in breast cancer cells than in MCF10A cells (Figure 1G). These results suggested that TPD52 was upregulated in breast cancer cell lines, with the MDA-MB-231 cell line showing the highest expression level and the BT474 cell line exhibiting the lowest expression level. Consequently, the MDA-MB-231 and BT474 cell lines were selected for further experimentation.
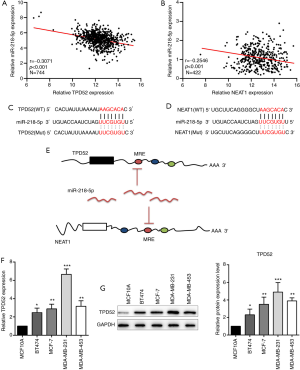
TPD52 promotes the growth and migration of breast cancer cells
A TPD52 expression vector or empty vector was transfected into BT474 cells, while TPD52 was knocked down in MDA-MB-231 cells via siRNA transfection (Figure S1). Western blotting revealed that knockdown of TPD52 inhibited Ki-67, MCM2, and PCNA expression at the protein level (Figure 2A). EdU and colony formation assays revealed that MDA-MB-231 cell proliferation was significantly inhibited by si-TPD52 transfection compared with si-NC transfection. The opposite effects were observed in the BT474 cell line with TPD52 overexpression; specifically, increased proliferation marker protein expression, cell proliferation, and colony formation, (Figure 2B,C,D). Overexpression of TPD52 promoted BT474 cell migration, whereas MDA-MB-231 cell migration was suppressed by TPD52 interference (Figure 2E,F,G,H). These data suggest that TPD52 is an oncogenic mRNA which promotes breast cancer cell proliferation.
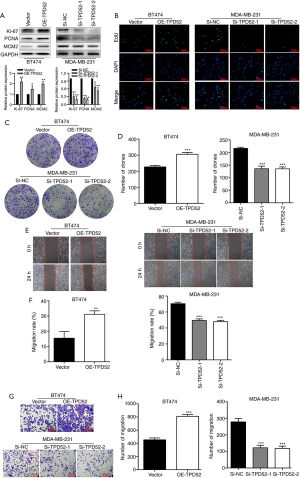
miR-218-5p regulates TPD52 and NEAT1
As revealed by RT-qPCR, the messenger RNA (mRNA) expression level of miR-218-5p in MDA-MB-231 cells was lower than that in BT474 cells, whereas NEAT1 expression was higher in MDA-MB-231 cells than in BT474 cells (Figure 3A,B). miR-218-5p inhibitors or mimics were transfected into the BT474 and MDA-MB-231 cell lines (transfection efficiency shown in Figure S2), and further RT-qPCR analysis revealed that miR-218-5p negatively regulated the expression levels of TPD52 and NEAT1 (Figure 3C). The transfection efficiency with a NEAT1 overexpression vector and NEAT1 siRNAs was measured by RT-qPCR. NEAT1 mRNA expression was decreased after transfection with NEAT1 siRNAs but increased after transfection with NEAT1 overexpression vector (Figure S3). Following NEAT1 overexpression or knockdown in the breast cancer cell lines, the RT-qPCR assay results showed that the downregulation of NEAT1 increased the mRNA expression levels of miR-218-5p and reduced those of TPD52, while the upregulation of NEAT1 had the opposite effects (Figure 3D,E). The sequences of lncRNA NEAT1 (chromosome 11, 65203019-65204039) and the 3'UTR region of TPD52 (chromosome 11, 80948905-80949911) were cloned into the luciferase reporter gene vector. Pattern diagrams showing the detailed binding sites between miR-218-5p/NEAT1 and miR-218-5p/TPD52 are displayed in Figure 1C,D. Overexpression of miR-218-5p reduced luciferase activity in the TPD52 and NEAT1 wild-type groups, but not in the mutant groups (Figure 3F,G). As shown in Figure 3H,I,J,K, the mRNA expression of TPD52 was elevated by NEAT1 overexpression or miR-218-5p silencing, and diminished by NEAT1 silencing or miR-218-5p overexpression. TPD52 protein expression was also increased by NEAT1 overexpression and decreased by miR-218-5p overexpression; the protein expression changes of TPD52 were similar to those of TPD52 mRNA (Figure 3L). To sum up, NEAT1 positively regulated the expression level of TPD52, and miR-218-5p mimics or inhibitors decreased the effects of NEAT1 overexpression or silencing, respectively, on TPD52 expression in the BT474 and MDA-MB-231 cell lines. These results confirmed that NEAT1 promotes the expression level of TPD52 in breast cancer cells by inhibiting miR-218-5p.
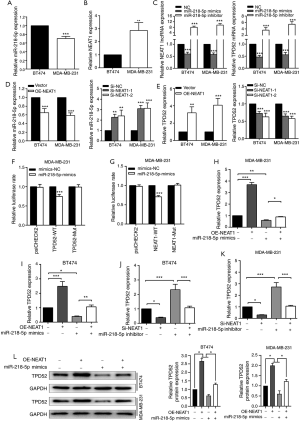
The NEAT1/miR-218-5p/TPD52 axis promotes the proliferation of breast cancer cells
The EdU and colony formation assay results (Figure 4) demonstrated that TPD52 overexpression partially rescued the inhibitory effects of miR-218-5p mimics or NEAT1 knockdown on breast cancer cell proliferation (Figure 4A,C). Meanwhile, TPD52 knockdown partially rescued the stimulatory effects of miR-218-5p inhibitor or NEAT1 upregulation on breast cancer cell proliferation (Figure 4B,D). These findings indicated that NEAT1 acted as an oncogene by targeting the miR-218-5p/TPD52 axis.
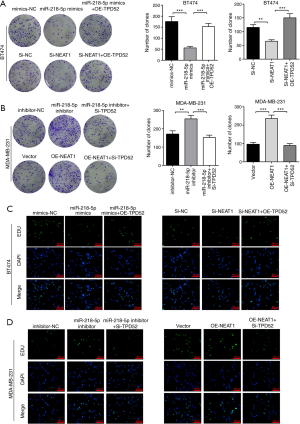
The NEAT1 miR-218-5p/TPD52 axis promotes the migration of breast cancer cells
Wound healing and Transwell assays (Figure 5) demonstrated that TPD52 overexpression partially rescued the negative effects of miR-218-5p mimics or NEAT1 downregulation on BT474 breast cancer cell migration (Figure 5A,C,E), while TPD52 knockdown partially rescued the stimulatory effects of miR-218-5p inhibitor or NEAT1 upregulation on breast cancer cell migration (Figure 5B,D,F). These results suggested that the stimulatory effects of NEAT1 were partially mediated by the miR-218-5p/TPD52 axis.
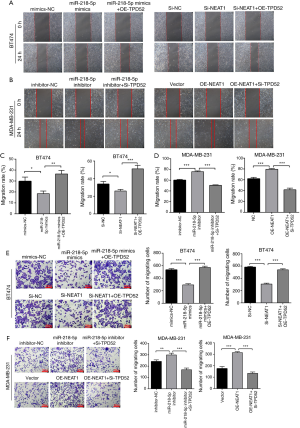
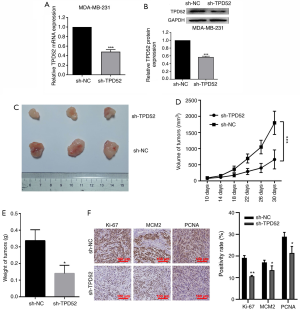
Downregulation of TPD52 suppresses tumor growth of breast cancer in vivo
To study the effects of TPD52 on breast cancer in vivo, MDA-MB-231 cells with stable knockdown of TPD52 were subcutaneously transplanted into nude mice. The mRNA and protein expression levels of TPD52 were decreased by sh-TPD52 transfection compared with sh-NC transfection (Figure 6A,B). The tumors of the mice were measured; the largest tumor had a volume of 1,864.9 mm3 and a longest diameter of 16.5 mm. Tumor growth was significantly inhibited in mice with TPD52 inhibition compared with the sh-NC group (Figure 6C,D). The average tumor weights were also compared between the groups, and the tumor weight in the TPD52 knockdown group was lower than that in the control group (Figure 6E). Immunohistochemical examination showed that the positive expression levels of Ki-67, MCM2, and PCNA in tumors derived from the TPD52 gene knockout group were reduced compared to those in the sh-NC group (Figure 6F). Thus, TPD52 knockdown inhibited breast cancer growth in vivo.
Discussion
TPD52 is a small coiled-coil motif-bearing protein belonging to the TPD52-like protein family, which is conserved from lower organisms to humans (9). Multiple studies have confirmed that the mRNA and protein expression levels of TPD52 show abnormal increases in a variety of malignancies, including prostate (18), breast (19), and colorectal (20) cancer. In a study of breast cancer, miR-34a inhibited the proliferation of breast cancer cells through its targeting of TPD52 (21). In the present study, we observed that TPD52 was upregulated in breast cancer cell lines compared with a normal breast cell line. The effects of modulating TPD52 expression on the breast cancer cell lines were also determined. It was found that the proliferation and migration of breast cancer cells were promoted in vitro, and their proliferation were promoted in vivo .
miRNAs are crucial regulators of mRNA expression and suppress the translation of complimentary mRNAs by binding to their 3'-UTRs. The expression levels of TPD52 and miR-218-5p, which is a tumor suppressor (22,23), were found to be negatively correlated in patients with breast cancer. Furthermore, bioinformatics analysis and a dual-luciferase reporter assay confirmed that miR-218-5p directly binds with NEAT1. Further experimentation revealed that the inhibition of miR-218-5p promoted the proliferation and migration of breast cancer cells, and that TPD52 knockdown partially rescued the stimulatory effects of the miR-218-5p inhibitor on breast cancer cell proliferation and migration. These results demonstrate that miR-218-5p can inhibit the proliferation and migration of breast cancer cells by targeting TPD52 .
It has been hypothesized that lncRNAs play an important role in cancer. For instance, a meta-analysis showed that the lncRNA NEAT1 is upregulated in various cancers, affects the prognosis, and is associated with a low overall survival time (24). Another study reported that NEAT1 epigenetically reduced E-cadherin expression by interacting with the G9a-DNMT1-Snail complex (25). In the present study, the upregulation of NEAT1 was found to promote the proliferation and migration of breast cancer cells. The ceRNA hypothesis suggests that RNAs are able to crosstalk by binding to miRNAs via the miRNA response elements in their 3'-UTRs, thereby preventing the miRNAs from binding to their target mRNAs (26). For instance, NEAT1 has been demonstrated to act as a ceRNA and positively promotes the expression level of EZH2 by sponging miR-101 in breast cancer cells (27). Several studies have indicated that NEAT1 induces breast cancer progression, is localized primarily in the nucleus, and has associations with various miRNAs, including miR-218 (28), miR-548 (29), and miR-448 (6). This study showed that NEAT1 upregulated the expression of TPD52 by competitively sponging miR-218-5p, thereby promoting the proliferation and migration of breast cancer cells.
In conclusion, this study has demonstrated that upregulation of TPD52 promotes cell proliferation and migration in breast cancer cell lines. NEAT1 can upregulate the expression levels of TPD52 in breast cancer cells by sponging miR-218-5p, thereby preventing miR-218-5p from inhibiting TPD52 expression by binding to its 3'-UTR. The upregulation of TPD52 by NEAT1 overexpression has also been shown to enhance cell proliferation and migration. The present study has provided a deeper understanding of the functions of the NEAT1/miR-218-5p/TPD52 axis in the pathophysiology of breast cancer, and our observations suggest that the lncRNA NEAT1 may be a promising biomarker and therapeutic target for human breast cancer.
Acknowledgments
Funding: This study was supported by the National Nature Science Foundation of China (grant Nos. 81601702, 81671931, 81772101, 81701920, 81801933, 81801932, 81871573, 81901976, 81901975, and 81971852), the Medical Scientific Research Foundation of Guangdong Province of China (grant No. A2020542, A2021446), and the Administrator Foundation of Nanfang Hospital (grant No. 2019B021).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-2668
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-2668
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-2668). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. d. All procedures performed using animals were approved by the Animal Experiments Committee of the Southern Medical University (approval Nos. L2018015 and 201690525). All applicable international, national and/or institutional guidelines for the care and use of animals were followed.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci 2017;108:1927-33. [Crossref] [PubMed]
- Qiao K, Ning S, Wan L, et al. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J Exp Clin Cancer Res 2019;38:418. [Crossref] [PubMed]
- Zhang M, Wu WB, Wang ZW, et al. lncRNA NEAT1 is closely related with progression of breast cancer via promoting proliferation and EMT. Eur Rev Med Pharmacol Sci 2017;21:1020-6. [PubMed]
- Jiang X, Zhou Y, Sun AJ. NEAT1 contributes to breast cancer progression through modulating miR-448 and ZEB1. J Cell Physiol 2018;233:8558-66. [Crossref] [PubMed]
- Kotapalli SS, Dasari C, Duscharla D, et al. All-Trans-Retinoic Acid Stimulates Overexpression of Tumor Protein D52 (TPD52, Isoform 3) and Neuronal Differentiation of IMR-32 Cells. J Cell Biochem 2017;118:4358-69. [Crossref] [PubMed]
- Goto Y, Nishikawa R, Kojima S, et al. Tumour-suppressive microRNA-224 inhibits cancer cell migration and invasion via targeting oncogenic TPD52 in prostate cancer. FEBS Lett 2014;588:1973-82. [Crossref] [PubMed]
- Boutros R, Fanayan S, Shehata M, et al. The tumor protein D52 family:many pieces, many puzzles. Biochem Biophys Res Commun 2004;325:1115-21. [Crossref] [PubMed]
- Byrne JA, Frost S, Chen Y, et al. Tumor protein D52 (TPD52) and cancer-oncogene understudy or understudied oncogene? Tumour Biol 2014;35:7369-82. [Crossref] [PubMed]
- Yang M, Wang X, Jia J, et al. Tumor protein D52-like 2 contributes to proliferation of breast cancer cells. Cancer Biother Radiopharm 2015;30:1-7. [Crossref] [PubMed]
- Zhao Z, Liu H, Hou J, et al. Tumor Protein D52 (TPD52) Inhibits Growth and Metastasis in Renal Cell Carcinoma Cells Through the PI3K/Akt Signaling Pathway. Oncol Res 2017;25:773-9. [Crossref] [PubMed]
- Zhang E, Han L, Yin D, et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res 2017;45:3086-101. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Pan Y, Li J, Zhang Y, et al. Slug-upregulated miR-221 promotes breast cancer progression through suppressing E-cadherin expression. Sci Rep 2016;6:25798. [Crossref] [PubMed]
- Liu Y, Chen C, Qian P, et al. Gd-metallofullerenol nanomaterial as non-toxic breast cancer stem cell-specific inhibitor. Nat Commun 2015;6:5988. [Crossref] [PubMed]
- Mao L, Zhan YY. ULK1 phosphorylates Exo70 to suppress breast cancer metastasis. Nat Commun 2020;11:117. [Crossref] [PubMed]
- Dasari C, Reddy KRK, Natani S, et al. Tumor protein D52 (isoform 3) interacts with and promotes peroxidase activity of Peroxiredoxin 1 in prostate cancer cells implicated in cell growth and migration. Biochim Biophys Acta Mol Cell Res 2019;1866:1298-309. [Crossref] [PubMed]
- Zhang Z, Wang J, Gao R, et al. Downregulation of MicroRNA-449 Promotes Migration and Invasion of Breast Cancer Cells by Targeting Tumor Protein D52 (TPD52). Oncol Res 2017;25:753-61. [Crossref] [PubMed]
- Li J, Li Y, Liu H, et al. The four-transmembrane protein MAL2 and tumor protein D52 (TPD52) are highly expressed in colorectal cancer and correlated with poor prognosis. PLoS One 2017;12:e0178515 [Crossref] [PubMed]
- Li G, Yao L, Zhang J, et al. Tumor-suppressive microRNA-34a inhibits breast cancer cell migration and invasion via targeting oncogenic TPD52. Tumour Biol 2016;37:7481-91. [Crossref] [PubMed]
- Litière S, Werutsky G, Fentiman IS, et al. Breast conserving therapy versus mastectomy for stage I-II breast cancer: 20 year follow-up of the EORTC 10801 phase 3 randomised trial. Lancet Oncol 2012;13:412-9. [Crossref] [PubMed]
- Hu Y, Xu K, Yagüe E. miR-218 targets survivin and regulates resistance to chemotherapeutics in breast cancer. Breast Cancer Res Treat 2015;151:269-80. [Crossref] [PubMed]
- Zhang Y, Lun L, Li H, et al. The Value of lncRNA NEAT1 as a Prognostic Factor for Survival of Cancer Outcome: A Meta-Analysis. Sci Rep 2017;7:13080. [Crossref] [PubMed]
- Li Y, Cheng C. Long noncoding RNA NEAT1 promotes the metastasis of osteosarcoma via interaction with the G9a-DNMT1-Snail complex. Am J Cancer Res 2018;8:81-90. [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Qian K, Liu G, Tang Z, et al. The long non-coding RNA NEAT1 interacted with miR-101 modulates breast cancer growth by targeting EZH2. Arch Biochem Biophys 2017;615:1-9. [Crossref] [PubMed]
- Zhao D, Zhang Y, Wang N, et al. NEAT1 negatively regulates miR-218 expression and promotes breast cancer progression. Cancer Biomark 2017;20:247-54. [Crossref] [PubMed]
- Ke H, Zhao L, Feng X, et al. NEAT1 is Required for Survival of Breast Cancer Cells Through FUS and miR-548. Gene Regul Syst Bio 2016;10:11-7. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


