
Lin28A promotes the proliferation and stemness of lung cancer cells via the activation of mitogen-activated protein kinase pathway dependent on microRNA let-7c
Introduction
Lung cancer ranks among the highest-incidence malignant tumors globally and is associated with high morbidity and fatality rates (1,2). Although molecular classification and targeted therapy of lung cancer has achieved significant progress in recent years, patients have limited benefit from regular therapies and the 5-year survival rate of the disease has not been effectively improved (3,4). Metastasis and recurrence are the leading causes of disease-related lung cancer deaths (2,5). Recently, the roles of cancer stem cells (CSCs) in the occurrence and development of tumors have received considerable attention (6-8). Lung cancer stem cell (LCSC) constitutes a highly carcinogenic cell subgroup, which exhibits similar biological characteristics as normal tissue stem cells, including an unlimited self-renewal ability, an extensive proliferative ability and a capacity to generate differentiated progeny (9,10). More and more evidences show that LCSCs play a crucial role in tumor progression, metastasis and drug resistance (11-13). Current anti-cancer therapies cannot eradicate LCSC clones. Instead, it facilitates to expand the LCSC pool and select for drug-resistant LCSC clones, leading to treatment resistance and subsequent recurrence in these patients (14-16).
However, currently, no large-scale clinical treatment targeting LCSCs exists, mainly owing to the challenges of LCSC isolation and purification. Moreover, the molecular mechanisms regulating LCSCs are still not clear. Therefore, the establishment of a culture method for LCSC enrichment and further illumination of the molecular mechanisms involved in the occurrence and development of LCSCs would be of great significance to the screening of more effective drugs to target LCSCs in lung cancer clinical research.
Since under 2-dimensional (2D) culture, cancer cells grow in a single layer and might differentiate and lose their stemness phenotypes rapidly (17,18), simulating the local tumor microenvironment in vitro using 2D culture method is challenging. Therefore, 2D culture is not suitable for mechanistic research and clinical transformation of LCSCs (19,20). Our previous work established a 3-dimensional (3D) culture system based on basement membrane extract-based (BME) with the addition of fibroblast growth factor 1 (FGF1) and insulin-like growth factor 1 (IGF1), which could provide the cultured cells with vital growth factors (21). Furthermore, the 3D structure facilitated the formation of tissue-specific structures to effectively mimic a local microenvironment in vivo. The 3D culture system we established could effectively achieve the enrichment and amplification of LCSCs. In our 3D culture system, FGF1 and IGF1 supported LCSC amplification and cancer stemness by activating mitogen-activated protein kinase (MAPK) signaling pathway. However, the molecular mechanisms of the amplification and development of LCSCs in our 3D culture system are still not entirely clear.
An increasing amount of research has evidenced microRNAs (miRNAs), as small non-coding RNAs responsible for the negative regulation of post-transcriptional gene expression (22-24), to be critical regulatory players in cancer cell stemness via their targeting of different genes (25-27). In lung cancer, miRNAs have been shown to be the chief molecular regulators involved in maintaining the metastatic, drug-resistant, and self-renewing properties of LCSCs through their effective targeting of vital genes involved in various signaling pathways (28,29). The combination of miRNAs with antitumor drugs has been associated with positive preclinical outcomes in multiple translational works (30-33). Therefore, miRNAs might serve as promising targets for eliminating CSCs.
Lin28A is one of the reprogramming factors to induce pluripotency in adult human fibroblast cells (34), which has been reported to be over-expressed in advanced human malignancies and play an important role in the maintenance of CSCs (35,36). Previous studies showed that miRNA let-7 binds to the 3’ UTR of Lin28A mRNA to negatively regulate Lin28A expression, while Lin28A itself blocks let-7 expression, thereby establishing a double negative feedback loop (36). However, none study focusing on the expression and roles of Lin28A in LCSCs has ever been reported.
In this study, to investigate the most critical miRNAs involved in the regulation of cancer cell stemness in our 3D culture system, we searched 2 popular miRNA databases and filtered out 25 candidate miRNAs which might have abnormal expression levels in lung cancer. We examined the levels of these 25 miRNAs in different lung cancer cell lines cultured in our 3D system. The levels of let-7 miRNAs, especially let-7c, were significantly down-regulated in LCSCs amplified in our 3D culture system. In previous studies, a reduced expression of let-7c was common in cancer and was usually correlated with a poor patient outcome (37-39). Overexpressing let-7 expression was found to inhibit sphere formation and carcinogenesis of CSCs, and reduce the proportion of undifferentiated cells; similarly, suppressing let-7 enhanced self-renewal and prompted aggressiveness (40-44). However, some studies have revealed that let-7c can also enhance tumor invasion and growth via the DVL3/β-catenin axis (45). Although let-7c is known to play an important dual role in regulating the tumorigenic and metastatic abilities of cancer cells, its role in LCSC development is still not very clear. Therefore, this study explored the functions of let-7c during the occurrence and development of lung cancer using LCSCs.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-2124).
Methods
Protocol registration
A protocol was prepared before the study with registration in 81702280 (the National Natural Science Foundation of China).
Patient information
A total of 125 patients with lung adenocarcinoma (LUAD; N=125 in total) who received partial lung resection in Tianjin Medical University Cancer Institute and Hospital’s Department of Lung Cancer. We also obtained 4 LUAD tissue specimens (N=4 in total), from which CD326+CD44+CD24− cells were isolated by flow sorting. None of the patients had received therapies, such as chemotherapy or radiotherapy, ahead of their operation. The Ethics Committee of Tianjin Medical University granted approval for this study (Approved No.: Ek2017055). Each experiment was carried out in adherence with the principles of the Declaration of Helsinki (as revised in 2013). Each patient gave written informed consent before participating in the study.
Cell lines and cell culture
The human LUAD cell lines A549 and H1299, and the lung squamous cell carcinoma (LUSC) cell lines NCI-H520 and SK-MES-1 (Chinese Academy of Medical Sciences’ tumor cell library) were subjected to short tandem repeat DNA profiling and mycoplasma testing. For the 2D culture system, A549, H1299, and NCI-H520 cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 Medium (Gibco BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Gibco BRL, Grand Island, NY, USA) at 37 °C under 5% CO2. SK-MES-1 was cultured in Minimal Essential Medium containing 10% fetal bovine serum, 1% non-essential amino acids, 1 mM sodium pyruvate, and 1% penicillin/streptomycin. For 3D culture, firstly, the addition of 50 µL/well Cultrex® Basement Membrane Extract (BME, R&D Systems, MN, USA) was made to 96-well plates, which were then kept at 37 °C for 1 hour, to form a reconstituted basement membrane with specific thickness and stiffness. Then, the BME-coated plate was seeded with suspended cells (1×104 cell/200 µL/well), which were examined at 3-day intervals. Cell aggregate was generated through migration of cells into the inner BME layer. At day 3 of culturing, the simultaneous addition of 20 ng/mL FGF1 and 50 ng/mL IGF1 was made to complete medium as described previously (21). In each experiment, we established 3 identical wells for each group of cells.
Lentivirus construction
For let-7c over-expression lentivirus construction, the full-length cDNA of human hsa-let-7c was amplified by PCR with forward primer and reverse primers containing EcoRI and BamHI sites, respectively. The primers sequence were FP:Ata gaa ttc GCA TCC GGG TTG AGG TAG TAG GTT GTA TGG TTT AGA GTT ACA CCC TGG GAG TTA; RP: Ata gga tcc AAA AAA TGC TCC AAG GAA AGC TAG AAG GTT GTA CAG TTA ACT CCC AGG GTG TAA C. Then the amplified let-7c fragment was inserted into pHBLV-CMV-MCS-EF1-ZsGreen-T2A-Puro lentiviral vectors (Hanbio Co., Ltd., Shanghai, China), and the constructed positive plasmid was confirmed by DNA sequencing. The recombinant lentivirus with let-7c sequence was generated by cotransfection in A549 cells as previously described (46). For Lin28A knockdown lentivirus construction, specific shRNA sequence based on the sequence of Lin28A (Gene ID: 79727, on NCBI) was designed. The synthesized Lin28A shRNA was inserted into plvx-U6-CMV-RFP-P2A-BSD lentiviral vectors. The lentivirus was generated as described above.
Mice and xenograft model construction
Female non-obese diabetic/severe combined immunodeficiency mice (7 weeks old; 17–18 g) were obtained from Beijing SPF Biotechnology Co., Ltd. (Beijing, China) and randomly divided into two groups. The animals were kept in a specific pathogen-free animal facility at Tianjin Medical University Cancer Hospital and Institute. We obtained different types of cells carrying constructed lentiviruses (A549-3D+F, A549OV-let-7c-3D+F, and A549sh-Lin28A-3D+F; N=5 in each group) and resuspended 5×104 cells in 100 µL of PBS with Matrigel, followed by administered the cells into the flanks of NOD-SCID mice via subcutaneous injection. All procedures involving animals received approval from the Ethics Committee for Animal Experiments at Tianjin Medical University Cancer Hospital and Institute (Approved No.: NSFC-AE-2020101) and adhered to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Flow cytometry analysis
Labelling of LCSCs (N=5 in each group) was performed using fluorescein isothiocyanate (FITC) anti-human CD326, APC anti-human CD44, and PE anti-human CD24 antibodies (BD Biosciences, San Jose, CA, USA). We selected isotype-matched immunoglobulin G1 antibodies (BD Biosciences, San Jose, CA, USA) to serve as a negative control. A BD FACS CantoTM II flow cytometer was employed for flow cytometry. The gating strategy used was described in our previous work (21).
Statistical analyses
Animals were excluded from a study for the reason of illness or mortality. And data points were excluded from analyses for the reason of biologically implausible values. Data were statistically analyzed with SPSS 20.0 and GraphPad Prism 5.0 software following the manufacturers’ instructions. Measurement data were expressed as means ± standard deviations. We analyzed correlations between 2 datasets using Spearman’s correlation coefficient. One- and 2-way analysis of variance with subsequent Bonferroni post-hoc tests was used for comparisons between 2 groups. Cumulative survival was determined via the Kaplan-Meier method. All data were normally distributed. P<0.05 was taken to indicate a statistically significant result.
More detailed methods can be found in the Supplemental data (Appendix 1).
Results
Lin28A/let-7c was dysregulated in LCSCs from the BME-based 3D culture system
In our previous studies, we used FGF1 and IGF1 to establish the conditional 3D culture based on BME with growth factors (3D+F) for LCSC amplification in A549 cells (A549-3D+F cells) in vitro (21). To investigate the most critical miRNA(s) involved in the regulation of stemness in A549-3D+F cells, we firstly filtered candidate miRNAs in online databases. We searched the Human miRNA Disease Database (HMDD, version 3.2) (47), in which experiment-supported evidence for human miRNA and disease associations is curated. We identified 305 miRNAs which are dysregulated in lung cancer, including 108 miRNAs that have direct causality with lung cancer. We also searched for miRNAs on the OncomiR database (48), and this search retrieved 379 miRNAs associated with lung cancer development, as well as 109 miRNAs associated with survival outcome of lung cancer. We obtained 25 differentially expressed miRNAs from the intersection of the 2 databases (Figure S1). Then, we collected the pellets of A549 cells from the 2D culture (A549-2D cells) and A549-3D+F cells, and performed quantitative-polymerase chain reaction (qPCR) to compare the RNA levels of the 25 differentially expressed miRNAs. In comparison with A549-2D cells, A549-3D+F cells exhibited dramatic increases in the levels of a number of miRNAs, such as miR-873, miR-31, miR-582, miR-196b, and miR-212. Meanwhile, some miRNAs were significantly decreased in A549-3D+F cells, including miR-99a, miR-145, miR-181c, miR-30a, miR-29b, let-7g, let-7f, miR-490, and let-7c. Among these miRNAs, let-7c, let-7f, and let-7g, which belong to the same miRNA family, had the lowest expression levels in A549-3D+F cells (Figure 1A).
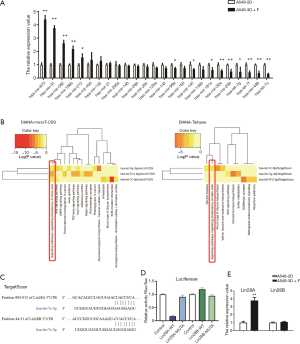
To investigate which biological processes let-7 miRNAs are involved in, we used DIANA-miRPath v2.0 (49), which is another popular database which can predict the pathways in which miRNA targets are enriched using the DIANA-microT-CDS algorithm and/or experimentally validated miRNA interactions derived from DIANA-TarBase v6.0. The results showed the let-7 miRNA target genes to be enriched in signaling pathways regulating the pluripotency of stem cells (Figure 1B), indicating that let-7 miRNAs are involved in the regulation of cancer stemness.
We then focused on the functions of let-7c, considering let-7c miRNAs had the lowest expression in A549-3D+F cells. We searched TargetScan to identify let-7c target genes (50), and found 1207 transcripts which are potentially targeted by let-7c. Among the top 10 target genes, Lin28A/B (Figure 1C), which is a reprogramming factor, has been observed to induce pluripotency in adult human fibroblast cells (51). The dual-luciferase reporter assay revealed that the relative luciferase activity of Lin28A 3’UTR, rather than Lin28B 3'UTR, was significantly decreased in HEK-293T cells transfected with let-7c mimic (Figure 1D). Then, we compared the transcription levels of Lin28A/B between A549-2D and A549-3D+F cells. Consistent with the results of the luciferase reporter assay, the transcription level of Lin28B showed no difference between the 2 groups, whereas the transcription level of Lin28A was significantly up-regulated in A549-3D+F cells (Figure 1E). All the above results indicated that Lin28A/let-7c was dysregulated in LCSCs from the BME-based 3D culture system, suggesting that Lin28A/let-7c might play roles in the development of LCSCs.
Lin28A/let-7c was expressed abnormally in LCSCs from other 3D cultured lung cancer cell lines
We further attempted to establish 3D+F culture to amplify LCSCs in another LUAD cell line, H1299. As before, we applied the co-stimulation of FGF1 and IGF1 in the BME-based 3D system to establish our 3D+F culture. In the spheroid formation assay, we could hardly observe apparent spheroids in H1299 cultured in 2D culture system (H1299-2D) (Figure 2A, left panel); however, several large spheroids were observed in H1299 cultured in 3D+F system (H1299-3D+F) (Figure 2A, right panel). The migration ability of H1299-2D and H1299-3D+F cells was evaluated via wound healing experiment. After 48 hours, the H1299-3D+F cell wound closure rate displayed a significant increase compared to that of H1299-2D cells (Figure 2B).
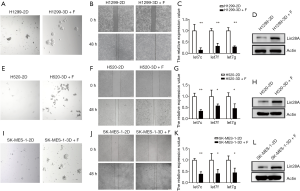
Further, we detected the levels of let-7 miRNAs and Lin28A in H1299-2D and H1299-3D+F cells. Consistent with the results in A549 cells, let-7c, let-7f, and let-7g were significantly down-regulated in H1299-3D+F cells compared to H1299-2D cells (Figure 2C), while the protein expression of Lin28A was significantly up-regulated in H1299-3D+F cells (Figure 2D), indicating that the expression loop of let-7c/Lin28A was largely dysregulated in LUAD CSCs.
We also used the 3D+F culture system to amplify LCSCs in the LUSC cell line H520. The sphere formation and wound healing assay results also revealed that cells cultured in the 3D+F system exhibited more stemness characteristics than those cultured in the 2D system (Figure 2E,F), implying that we had successfully and efficiently enriched and enhanced the stemness of LCSCs in both LUAD and LUSC cell lines in our 3D+F system. Furthermore, as expected, let-7 miRNAs showed down-regulation in cells cultured in the 3D+F system compared to those cultured in the 2D system (Figure 2G), while the protein expression levels of Lin28A were significantly up-regulated in cells cultured in the 3D+F system (Figure 2H). Besides that, we applied the 3D+F culture system to another LUSC cell line SK-MES-1, and observed the similar results in SK-MES-1 (Figure 2I,J,K,L). All the results above indicated that the abnormal expression of the let-7c/Lin28A loop was a common phenomenon in different subtypes of LCSCs.
Lin28A/let-7c was dysregulated in LCSCs from lung cancer tissues and its abnormal expression was correlated with poor survival outcomes
As CD326+, CD44+, CD24− has been reported as the characteristic phenotypic markers on the cell surface of LCSCs (52,53), CD326+CD44+CD24− cells were termed as the LCSCs in this research. Four clinical LUAD tissue samples were collected, from which CD326+CD44+CD24− cells were isolated (Figure 3A) by flow sorting. Then, we examined the RNA levels of let-7c, let-7f, and let-7g individually. Consistent with the results for LCSCs from the 3D culture system, the levels of let-7c were significantly decreased in the CD326+CD44+CD24− group compared with the non-CD326+CD44+CD24− group (Figure 3B). Furthermore, the expression levels of Lin28A were significantly up-regulated in CD326+CD44+CD24− cells, whereas the expression levels of Lin28B showed no difference between the 2 groups (Figure 3C).
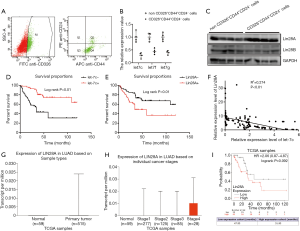
We then collected 125 LUAD tumor tissue specimens from Tianjin Medical University Cancer Institute and Hospital (Table 1) and explored let-7c/Lin28A’s association with patients’ overall survival (OS) by performing qPCR. The patients were stratified into 2 groups according to the transcription levels of either let-7c or Lin28A, following which their survival was analyzed by applying the Kaplan-Meier method. Low levels of let-7c, as well as a high expression of Lin28A, were found to be correlated with poor survival outcomes (Figure 3D,E). Furthermore, Spearman’s rank correlation revealed the levels of let-7c to be significantly negatively correlated with those of Lin28A in the clinical LUAD tumor samples (Figure 3F).

Full table
To further explore the effects of Lin28A in LUAD occurrence and development in a larger group of samples, we analyzed data from The Cancer Genome Atlas (TCGA). We found the level of Lin28A in LUAD primary tissue samples to show a significant increase compared to that in the matched adjacent non-tumor tissue samples (Figure 3G) (54). The analysis based on Lin28A expression in different cancer types showed the level of Lin28A in Stage 4 tissue samples to be markedly elevated compared to that in stage 1–3 tissue samples (Figure 3H) (54). Further, the TCGA survival analysis consistently evidenced a correlation between an increased level of Lin28A in lung cancer primary tissues and poor survival (Figure 3I) (55). Collectively, these results indicated that Lin28A/let-7c was dysregulated in LCSCs from lung cancer tissues and its abnormal expression was correlated with poor survival outcomes of LUAD.
Let-7c inhibits the maintenance of LCSC properties via disruption of MAPK signaling pathway
To probe the biological role of let-7c in LCSC enrichment and amplification, we induced let-7c over-expression in A549 cells, which were then cultured in the 3D+F system (A549OV-let-7c-3D+F). We examined the proportions of CD326+CD44+CD24− cells in A549-2D, A549-3D, A549-3D+F, and A549OV-let-7c-3D+F cells using flow cytometry. Consistent with our previous findings, an increase in the expression levels of CD326 was observed in A549-3D cells compared to A549-2D cells, and the increase was more dramatic in A549-3D+F cells (Figure 4A). However, fewer CD326+ cells were observed among the A549OV-let-7c-3D+F cells compared to the A549-3D+F cells. Next, CD326+ cells were gated, and the proportion of CD44+CD24− cells in the CD326+ population was analyzed. The proportion of CD326+CD44+CD24− cells among A549-3D+F cells was larger than that among A549-2D and A549-3D cells (Figure 4A). Moreover, fewer CD326+CD44+CD24− cells were observed among A549OV-let-7c-3D+F cells than among A549-3D+F cells, indicating that let-7c inhibited the expression levels of LCSC phenotypic markers.
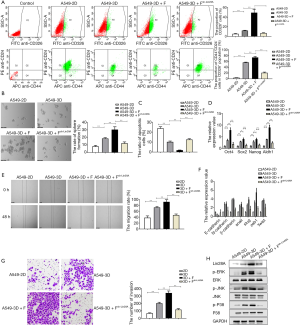
As LCSCs are considered to possess the characteristic of high self-renewal, we further compared the spheroid formation capacities of A549-2D, A549-3D, A549-3D+F, and A549OV-let-7c-3D+F cells to examine the impact of let-7c on LCSC stemness in these cells. A549-3D+F cells formed larger and more numerous colonies compared to A549-2D and A549-3D cells, but fewer and smaller cell colonies were detected in A549OV-let-7c-3D+F cells compared to A549-3D+F cells (Figure 4B). Next, RT-qPCR was performed for the detection of the mRNA expressions of classical stemness genes (Oct4, Sox2, Nanog, and Aldh1). Each gene showed significant up-regulation at the mRNA level in A549-3D+F cells in comparison with A549-2D and A549-3D cells, whereas significant mRNA down-regulation of the genes was observed in A549OV-let-7c-3D+F cells in comparison with A549-3D+F cells (Figure 4C). These results indicated that A549OV-let-7c-3D+F cells displayed significantly lower expression of phenotypic and genetic LCSC markers, suggesting that let-7c might be an inhibitor of LCSC enrichment and proliferation.
As LCSCs are considered to be more resistant to starvation-induced apoptosis and have more strong resistance to chemotherapy drugs (52,56). Annexin-V apoptosis assay was then conducted to examine cell apoptosis among A549-2D, A549-3D, A549-3D+F, and A549OV-let-7c-3D+F cells. The flow cytometry analysis showed less apoptotic cells in A549-3D+F compared to either A549-2D or A549-3D cells, but more apoptotic cells were detected in A549OV-let-7c-3D+F compared to A549-3D+F cells (Figure 4D, Figure S2A). Further, the drug resistance assay showed although A549-3D+F were more resistant to cisplatin treatment, A549OV-let-7c-3D+F were more sensitive to cisplatin treatment than A549-3D+F cells (Figure S2B).
Next, since LCSCs are considered to possess the characteristic of high migration and invasion abilities, we examined the migratory and invasive abilities of A549-2D, A549-3D, A549-3D+F, and A549OV-let-7c-3D+F cells by performing wound healing and Transwell invasion assays. After 48 hours, the wound closure rate of A549-3D+F cells was markedly increased compared to those of A549-2D and A549-3D cells (Figure 4E). With epithelial-mesenchymal transition (EMT) being the most critical cellular process in tumor invasion and spread (57), qPCR was carried out to detect the enriched transcription of biomarkers related to EMT. The expression of the epithelial cell biomarker E-cadherin, which is known for its conventional use, showed a decrease in A549-3D+F cells in comparison to A549-2D and A549-3D cells, although the levels of N-cadherin and β-catenin, which are well-documented mesenchymal biomarkers, and EMT-associated transcription factors (Snail, Slug, ZEB1, and Twist) displayed an increase. Meanwhile, the above-mentioned mesenchymal cell biomarkers were decreased significantly in A549OV-let-7c-3D+F cells (Figure 4F). Further, at 48 hours, the number of migratory cells observed to have crossed the Matrigel layer was elevated in the A549-3D+F group compared to in the A549-2D and A549-3D groups (Figure 4G). However, in the wound healing and Transwell invasion assays, we found that far fewer cells had migrated in the A549OV-let-7c-3D+F group compared to those in the A549-3D+F group (Figure 4E,G). All of these results suggested that the elevated migration and invasion abilities of A549-3D+F cells were reversed by over-expressing let-7c.
Since our previous observations had indicated that 3D+F culture had the effect of promoting LCSC amplification and stemness via MAPK signaling pathway (21), we then collected the supernatants of A549-2D, A549-3D, A549-3D+F, and A549OV-let-7c-3D+F cells, after which we examined the levels of phosphorylated p38, c-Jun N-terminal kinase (JNK), and extracellular regulated protein kinases (ERK) via western blot. A549-3D and A549-3D+F cells exhibited up-regulation of the phosphorylation levels of these 3 major subunits of MAPK protein family in comparison with A549-2D cells; however, the phosphorylation levels were obviously down-regulated in A549OV-let-7c-3D+F cells (Figure 4H), which implied that let-7c inhibited the maintenance of LCSC properties via disruption of MAPK signaling pathway.
Lin28A plays an indispensable role in tumorigenesis and the promotion of LCSC expansion via MAPK signaling pathway
Next, we transfected A549 cells with Lin28A short hairpin RNA (shRNA) and cultured them in the 3D+F system (A549sh-Lin28A-3D+F) to investigate the function of Lin28A in LCSC enrichment and proliferation. By flow cytometry analysis, fewer CD326+ cells and CD326+CD44+CD24− cells were observed among A549sh-Lin28A-3D+F cells than among A549-3D+F cells (Figure 5A), indicating Lin28A to be critical for the expression of phenotypic markers of LCSCs.
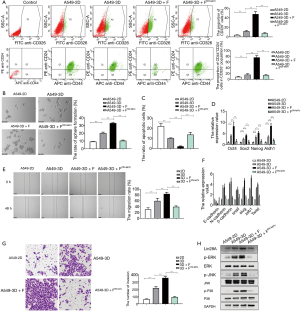
We further investigated the spheroid formation capacities of the cells, and observed the A549sh-Lin28A-3D+F group to have fewer and smaller cell colonies than the A549-3D+F group (Figure 5B). Furthermore, we observed the A549sh-Lin28A-3D+F group to have significant down-regulation of Oct4, Sox2, Nanog, and Aldh1 at the mRNA level in comparison with the A549-3D+F group (Figure 5C). Annexin-V apoptosis assay showed more apoptotic cells in A549sh-Lin28A-3D+F compared to A549-3D+F cells (Figure 5D, Figure S2C). And the drug resistance assay showed A549sh-Lin28A-3D+F were less resistant to cisplatin treatment than A549-3D+F cells (Figure S2D). These results indicated that A549sh-Lin28A-3D+F cells displayed significantly lower expression of LCSC phenotypic and genetic markers, even weakened the resistance to apoptosis and chemotherapy drugs, indicating that Lin28A is indispensable for the enrichment and proliferation of LCSCs.
The results of the wound healing assay showed the wound closure rate of A549sh-Lin28A-3D+F cells was significantly lower than that of A549-3D+F cells after 48 hours (Figure 5E). The results of qPCR showed E-cadherin to be increased, but N-cadherin, β-catenin, Snail, Slug, ZEB1, and Twist to be decreased in A549sh-Lin28A-3D+F cells in comparison with A549-3D+F cells (Figure 5F). Furthermore, the results of the Transwell invasion assay indicated that fewer A549sh-Lin28A-3D+F cells than A549-3D+F cells had migrated across the Matrigel layer (Figure 5G). These results suggested that the elevated migration and invasion abilities of A549-3D+F cells were also dependent on Lin28A functions.
Then, we examined the levels of phosphorylated p38, JNK, and ERK in the supernatant of A549-2D, A549-3D, A549-3D+F, and A549sh-Lin28A-3D+F cells, and observed that the phosphorylation levels were obviously down-regulated in the A549sh-Lin28A-3D+F group (Figure 5H). The results above implied that Lin28A plays an indispensable role in tumorigenesis and promotes LCSC expansion via MAPK signaling pathway.
Knockdown of Lin28A or over-expression of let-7c inhibited carcinogenesis and disrupted LCSC expansion in vivo
Subsequently, we set out to examine the effect of Lin28A on oncogenesis in vivo. A xenograft model was established by administering non-obese diabetic/severe combined immunodeficiency (NOD-SCID) mice with a subcutaneous injection of cells transfected with A549sh-Lin28A lentiviral constructs or their control cells. On day 24 after the injection, we observed a minimum 2-fold increase in the average tumor volumes in the A549sh-Lin28A group in comparison with the control (Figure 6A). We also investigated the roles of let-7c in tumorigenesis by injecting cells transduced with the A549OV-let-7c lentiviral constructs in a similar manner. Consistently, the growth rates of tumors in the A549OV-let-7c group far surpassed the rates in the H520OV−CTRL group (Figure 6B).
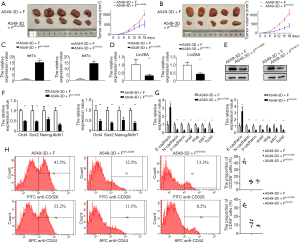
Additionally, the RNA levels of let-7c and protein levels of Lin28A in xenografts were examined. As expected, the qPCR analysis demonstrated that the transcription of let-7c was elevated in both the A549sh-Lin28A and A549OV-let-7c groups compared with their respective controls (Figure 6C). The qPCR and western blot results showed down-regulation of Lin28A in the A549sh-Lin28A and A549OV-let-7c groups (Figure 6D,E), as well as reduced mRNA levels of Oct4, Sox2, Nanog, and Aldh1 in the A549sh-Lin28A and A549OV-let-7c groups (Figure 6F). Similarly, the mRNA levels of E-cadherin were reduced, while those of N-cadherin, β-catenin, Snail, Slug, ZEB1, and Twist1 exhibited an increase (Figure 6G).
We further extracted the tumor tissues from the mice and analyzed the proportions of CSCs by flow cytometry. Markedly fewer CD326+ and CD44+ cells were observed in A549sh-Lin28A and A549OV-let-7c mice compared to A549 group (Figure 6H). All of these results implied that knockdown of Lin28A or over-expression of let-7c could inhibit carcinogenesis and disrupt LCSC expansion in vivo.
Discussion
The dysregulation of miRNA expression is related to and is functionally important in many diseases, including cancer (23,28). Among all tumor-suppressing miRNAs, a decrease in let-7 is highly common in tumors and is usually associated with a poor prognostic outcome (36,58,59). Functionally, the reduction of let-7 miRNAs leads to over-expression of their carcinogenic targets, such as resistance to audiogenic seizures (RAS), MYC proto-oncogene (MYC), and high mobility group AT-hook 2 (HMGA2) (60-62). Furthermore, in many cancers and embryonic stem cells, although the primary transcripts of let-7 are still actively transcribed, mature let-7 is not produced as expected, due to the 2 highly conserved RNA-binding proteins Lin28A and Lin28B directly bind to pre-let-7 or pri-let-7 to inhibit the biogenesis of mature let-7 miRNAs (36). In particular, binding of Lin28A/B to the terminal loop of the precursor let-7 results in the recruitment of the terminal uridylyl transferase ZCCHC11, which polyuridylates pre-let-7, thus blocking miRNA biogenesis and tumor-suppressing functions (36). Lin28A/B blocks the expression of let-7, which negatively regulates Lin28A/B levels through its binding to the 3’UTR of Lin28A/B mRNA; through this, a dual negative feedback loop is established (63).
Lin28A has been shown as a stem cell pluripotency factor (51). Treatment of adult fibroblasts with Oct4, Sox2, Nanog and Lin28A allowed for their successful reprogramming to induced pluripotent stem cells (PSCs) (64). The up-regulation and activation of Lin28A/B is a hallmark of in multiple malignancies, including glioblastoma, and cancers of the ovaries, stomach, prostate, and breast (34,35). Although the functions and mechanisms of the double-negative feedback loop of Lin28/let-7 in CSCs have been elucidated in many cancers, the relationship and role of Lin28/let-7 in LCSCs are still not well elucidated.
In this study, we detected that the levels of let-7 miRNAs, and found that, in particular, let-7c was significantly down-regulated in LCSCs amplified in our 3D culture system. Let-7c is a member of the let-7 family which is commonly known as a putative tumor suppressor of several cancer cell lines (37,38,40), but is also reported to play an important dual role in regulating the tumorigenic and metastatic abilities of cancer cells (45). Further, let-7c can target several genes, such as homeobox A1 (HOXA1), integrin subunit beta 3 (ITGB3), mitogen-activated protein kinase kinase kinase kinase 3 (MAP4K3), and tribbles pseudokinase 2 (TRIB2), to inhibit non-small cell lung cancer proliferation, migration, and tumorigenesis (65-67). Regarding its roles in regulating stem cells, let-7c has been identified to significantly abolish symmetric division of stem cells by inhibiting Wnt signaling (68).
However, there is no direct evidence of the relationship between let-7c and Lin28A/B in lung cancer. Our search for let-7c target genes on TargetScan showed that Lin28B was among the top 10 genes. Unexpectedly, the transcription and protein levels of Lin28B showed no difference between LCSCs and non-LCSCs, whereas the expression of Lin28A was significantly up-regulated in LCSCs amplified in our 3D culture system. The results indicated that although Lin28A and Lin28B have both been reported to play roles in the progression of lung cancer, Lin28A appeared to be more important to the enrichment and amplification of LCSCs.
The abnormal expression of Lin28A/let-7c was not only detected in LCSCs from A549 cells, but also in LCSCs from other lung cancer cell lines, including LUAD and LUSC cell lines. We also uncovered that Lin28A/let-7c was dysregulated in LCSCs obtained from lung cancer tissues. Furthermore, the abnormal expression of Lin28A/let-7c was correlated with poor survival outcomes in patients from our cohort and the TCGA database. Let-7c over-expression or inhibition of Lin28A suppressed stem cell self-renewal and carcinogenesis, and reduced the proportion of undifferentiated cells. All of these data imply that Lin28A and let-7c, but not Lin28B or other let-7 miRNAs, might play vital roles in the development of LCSCs in different subtypes of lung cancer.
Hedgehog, Wnt, and Notch are the canonical signaling pathways involved in mediating various stem cell characteristics, such as self-renewal, cell fate decisions, survival, proliferation, and differentiation (69,70). However, in our previous studies, we demonstrated that the growth factors promoting LCSC amplification and stemness mainly depended on MAPK signaling pathway (21). Lin28A/let-7c was also found to regulate the properties of LCSCs via MAPK signaling pathway. Ras genes are regulated by let-7 miRNAs due to the 3’-UTRs of these genes containing let-7 complementary sites (60). Deletion of let-7c has been reported to promote the activation of the JNK and P38 MAPK pathways in mesenchymal stem cells (41). Tsanov et al. found that Lin28A was phosphorylated by MAPK/ERK in PSCs, which increased its levels via post-translational stabilization (71). Our findings inversely indicated that MAPK signaling was the downstream pathway of Lin28A.
In this study, we have established an efficient 3D culture method for the amplification of different subtypes of LCSCs, with the intention of overcoming the challenges faced in the purification and amplification of LCSCs which limit large-scale clinical treatment targeting LCSCs. We have also illuminated the molecular mechanisms of Lin28A/let-7c/MAPK signaling pathway involved in the proliferation and development of LCSCs, even defining an axis that connects the gene, miRNA, signaling pathway, and stem cell properties. This pathway might be a potential therapeutic target for reducing and even eliminating LCSCs in clinical research in the future.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (grant No. 82072588, 81702280, and 81872143), the National Science and Technology Support Program of China (grant No. 2018ZX09201015), the National Key Research and Development Program of China: The Net construction of human genetic resource Bio-bank in North China (grant No. 2016YFC1201703), the Project of Science and Technology of Tianjin (grant No. 18JCQNJC82700), and the Key Project of Tianjin Health and Family Planning Commission (grant No. 16KG126).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-2124
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-2124
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-2124). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of Tianjin Medical University (Approved No.: Ek2017055). All experiments were performed in accordance with the principles of the Declaration of Helsinki (as revised in 2013) and written consent was obtained from all patients. All procedures involving animals were approved by the Ethics Committee for Animal Experiments at the Tianjin Medical University Cancer Hospital and Institute (Approved No.: NSFC-AE-2020101) and were performed in accordance with the Guide for the Care and Use of Laboratory Animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Yang CY, Yang JC, Yang PC. Precision Management of Advanced Non-Small Cell Lung Cancer. Annu Rev Med 2020;71:117-36. [Crossref] [PubMed]
- Gridelli C, Rossi A, Carbone DP, et al. Non-small-cell lung cancer. Nat Rev Dis Primers 2015;1:15009. [Crossref] [PubMed]
- Kim C, Giaccone G. Precision oncology in non-small-cell lung cancer: opportunities and challenges. Nat Rev Clin Oncol 2018;15:348-9. [Crossref] [PubMed]
- Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018;29:iv192-237. [Crossref]
- Greten FR. Cancer: Tumour stem-cell surprises. Nature 2017;543:626-7. [Crossref] [PubMed]
- Batlle E, Clevers H. Cancer stem cells revisited. Nat Med 2017;23:1124-34. [Crossref] [PubMed]
- Beck B, Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer 2013;13:727-38. [Crossref] [PubMed]
- Koren A, Motaln H, Cufer T. Lung cancer stem cells: a biological and clinical perspective. Cell Oncol (Dordr) 2013;36:265-75. [Crossref] [PubMed]
- Moharil RB, Dive A, Khandekar S, et al. Cancer stem cells: An insight. J Oral Maxillofac Pathol 2017;21:463. [Crossref] [PubMed]
- MacDonagh L, Gray SG, Breen E, et al. Lung cancer stem cells: The root of resistance. Cancer Lett 2016;372:147-56. [Crossref] [PubMed]
- Najafi M, Farhood B, Mortezaee K. Cancer stem cells (CSCs) in cancer progression and therapy. J Cell Physiol 2019;234:8381-95. [Crossref] [PubMed]
- Heng WS, Gosens R, Kruyt FAE. Lung cancer stem cells: origin, features, maintenance mechanisms and therapeutic targeting. Biochem Pharmacol 2019;160:121-33. [Crossref] [PubMed]
- Atashzar MR, Baharlou R, Karami J, et al. Cancer stem cells: A review from origin to therapeutic implications. J Cell Physiol 2020;235:790-803. [Crossref] [PubMed]
- Ayob AZ, Ramasamy TS. Cancer stem cells as key drivers of tumour progression. J Biomed Sci 2018;25:20. [Crossref] [PubMed]
- Bajaj J, Diaz E, Reya T. Stem cells in cancer initiation and progression. J Cell Biol 2020;219:e201911053 [Crossref] [PubMed]
- Sreepadmanabh M, Toley BJ. Investigations into the cancer stem cell niche using in-vitro 3-D tumor models and microfluidics. Biotechnol Adv 2018;36:1094-110. [Crossref] [PubMed]
- Pereira LP, Silva P, Duarte M, et al. Targeting Colorectal Cancer Proliferation, Stemness and Metastatic Potential Using Brassicaceae Extracts Enriched in Isothiocyanates: A 3D Cell Model-Based Study. Nutrients 2017;9:368. [Crossref] [PubMed]
- Bielecka ZF, Maliszewska-Olejniczak K, Safir IJ, et al. Three-dimensional cell culture model utilization in cancer stem cell research. Biol Rev Camb Philos Soc 2017;92:1505-20. [Crossref] [PubMed]
- Paolillo M, Colombo R, Serra M, et al. Stem-Like Cancer Cells in a Dynamic 3D Culture System: A Model to Study Metastatic Cell Adhesion and Anti-Cancer Drugs. Cells 2019;8:1434. [Crossref] [PubMed]
- Liu P, Zhang R, Yu W, et al. FGF1 and IGF1-conditioned 3D culture system promoted the amplification and cancer stemness of lung cancer cells. Biomaterials 2017;149:63-76. [Crossref] [PubMed]
- Cheng CJ, Bahal R, Babar IA, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature 2015;518:107-10. [Crossref] [PubMed]
- Rusek AM, Abba M, Eljaszewicz A, et al. MicroRNA modulators of epigenetic regulation, the tumor microenvironment and the immune system in lung cancer. Mol Cancer 2015;14:34. [Crossref] [PubMed]
- Khan AQ, Ahmed EI, Elareer NR, et al. Role of miRNA-Regulated Cancer Stem Cells in the Pathogenesis of Human Malignancies. Cells 2019;8:840. [Crossref] [PubMed]
- Agliano A, Calvo A, Box C. The challenge of targeting cancer stem cells to halt metastasis. Semin Cancer Biol 2017;44:25-42. [Crossref] [PubMed]
- Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 2015;15:321-33. [Crossref] [PubMed]
- Yan H, Bu P. Non-coding RNAs in cancer stem cells. Cancer Lett 2018;421:121-6. [Crossref] [PubMed]
- Xue J, Yang J, Luo M, et al. MicroRNA-targeted therapeutics for lung cancer treatment. Expert Opin Drug Discov 2017;12:141-57. [Crossref] [PubMed]
- Garofalo M, Croce CM. Role of microRNAs in maintaining cancer stem cells. Adv Drug Deliv Rev 2015;81:53-61. [Crossref] [PubMed]
- Van Roosbroeck K, Fanini F, Setoyama T, et al. Combining Anti-Mir-155 with Chemotherapy for the Treatment of Lung Cancers. Clin Cancer Res 2017;23:2891-904. [Crossref] [PubMed]
- Jeong K, Yu YJ, You JY, et al. Exosome-mediated microRNA-497 delivery for anti-cancer therapy in a microfluidic 3D lung cancer model. Lab Chip 2020;20:548-57. [Crossref] [PubMed]
- Lee J, Choi KJ, Moon SU, et al. Theragnosis-based combined cancer therapy using doxorubicin-conjugated microRNA-221 molecular beacon. Biomaterials 2016;74:109-18. [Crossref] [PubMed]
- Boca S, Gulei D, Zimta AA, et al. Nanoscale delivery systems for microRNAs in cancer therapy. Cell Mol Life Sci 2020;77:1059-86. [Crossref] [PubMed]
- Carmel-Gross I, Bollag N, Armon L, et al. LIN28: A Stem Cell Factor with a Key Role in Pediatric Tumor Formation. Stem Cells Dev 2016;25:367-77. [Crossref] [PubMed]
- Wang T, Wang G, Hao D, et al. Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer. Mol Cancer 2015;14:125. [Crossref] [PubMed]
- Balzeau J, Menezes MR, Cao S, et al. The LIN28/let-7 Pathway in Cancer. Front Genet 2017;8:31. [Crossref] [PubMed]
- Mulholland EJ, Green WP, Buckley NE, et al. Exploring the Potential of MicroRNA Let-7c as a Therapeutic for Prostate Cancer. Mol Ther Nucleic Acids 2019;18:927-37. [Crossref] [PubMed]
- Tang H, Ma M, Dai J, et al. miR-let-7b and miR-let-7c suppress tumourigenesis of human mucosal melanoma and enhance the sensitivity to chemotherapy. J Exp Clin Cancer Res 2019;38:212. [Crossref] [PubMed]
- Jilek JL, Zhang QY, Tu MJ, et al. Bioengineered Let-7c Inhibits Orthotopic Hepatocellular Carcinoma and Improves Overall Survival with Minimal Immunogenicity. Mol Ther Nucleic Acids 2019;14:498-508. [Crossref] [PubMed]
- Huang M, Gong X. Let-7c Inhibits the Proliferation, Invasion, and Migration of Glioma Cells via Targeting E2F5. Oncol Res 2018;26:1103-11. [Crossref] [PubMed]
- Liu GX, Ma S, Li Y, et al. Hsa-let-7c controls the committed differentiation of IGF-1-treated mesenchymal stem cells derived from dental pulps by targeting IGF-1R via the MAPK pathways. Exp Mol Med 2018;50:1-14. [Crossref] [PubMed]
- Fu X, Mao X, Wang Y, et al. Let-7c-5p inhibits cell proliferation and induces cell apoptosis by targeting ERCC6 in breast cancer. Oncol Rep 2017;38:1851-6. [Crossref] [PubMed]
- Guo Y, Li S, Qu J, et al. Let-7c inhibits metastatic ability of mouse hepatocarcinoma cells via targeting mannoside acetylglucosaminyltransferase 4 isoenzyme A. Int J Biochem Cell Biol 2014;53:1-8. [Crossref] [PubMed]
- Sun D, Layer R, Mueller AC, et al. Regulation of several androgen-induced genes through the repression of the miR-99a/let-7c/miR-125b-2 miRNA cluster in prostate cancer cells. Oncogene 2014;33:1448-57. [Crossref] [PubMed]
- Xie Y, Zhang H, Guo XJ, et al. Let-7c inhibits cholangiocarcinoma growth but promotes tumor cell invasion and growth at extrahepatic sites. Cell Death Dis 2018;9:249. [Crossref] [PubMed]
- Zhang R, Zhang F, Sun Z, et al. LINE-1 Retrotransposition Promotes the Development and Progression of Lung Squamous Cell Carcinoma by Disrupting the Tumor-Suppressor Gene FGGY. Cancer Res 2019;79:4453-65. [Crossref] [PubMed]
- Huang Z, Shi J, Gao Y, et al. HMDD v3.0: a database for experimentally supported human microRNA-disease associations. Nucleic Acids Res 2019;47:D1013-7. [Crossref] [PubMed]
- Wong NW, Chen Y, Chen S, et al. OncomiR: an online resource for exploring pan-cancer microRNA dysregulation. Bioinformatics 2018;34:713-5. [Crossref] [PubMed]
- Paraskevopoulou MD, Vlachos IS, Hatzigeorgiou AG. DIANA-TarBase and DIANA Suite Tools: Studying Experimentally Supported microRNA Targets. Curr Protoc Bioinformatics 2016;55:12.14.1-12.14.18.
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4:e05005 [Crossref] [PubMed]
- Zhang J, Ratanasirintrawoot S, Chandrasekaran S, et al. LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell Stem Cell 2016;19:66-80. [Crossref] [PubMed]
- Leon G, MacDonagh L, Finn SP, et al. Cancer stem cells in drug resistant lung cancer: Targeting cell surface markers and signaling pathways. Pharmacol Ther 2016;158:71-90. [Crossref] [PubMed]
- Eramo A, Haas TL, De Maria R. Lung cancer stem cells: tools and targets to fight lung cancer. Oncogene 2010;29:4625-35. [Crossref] [PubMed]
- Yan Z, Wang Q, Lu Z, et al. OSluca: An Interactive Web Server to Evaluate Prognostic Biomarkers for Lung Cancer. Front Genet 2020;11:420. [Crossref] [PubMed]
- Győrffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 2013;8:e82241 [Crossref] [PubMed]
- Wang L, Liu X, Ren Y, et al. Cisplatin-enriching cancer stem cells confer multidrug resistance in non-small cell lung cancer via enhancing TRIB1/HDAC activity. Cell Death Dis 2017;8:e2746 [Crossref] [PubMed]
- Mittal V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu Rev Pathol 2018;13:395-412. [Crossref] [PubMed]
- Lee H, Han S, Kwon CS, et al. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2016;7:100-13. [Crossref] [PubMed]
- Sun X, Liu J, Xu C, et al. The insights of Let-7 miRNAs in oncogenesis and stem cell potency. J Cell Mol Med 2016;20:1779-88. [Crossref] [PubMed]
- Oliveira-Mateos C, Sánchez-Castillo A, Soler M, et al. The transcribed pseudogene RPSAP52 enhances the oncofetal HMGA2-IGF2BP2-RAS axis through LIN28B-dependent and independent let-7 inhibition. Nat Commun 2019;10:3979. [Crossref] [PubMed]
- Gunzburg MJ, Sivakumaran A, Pendini NR, et al. Cooperative interplay of let-7 mimic and HuR with MYC RNA. Cell Cycle 2015;14:2729-33. [Crossref] [PubMed]
- Guo L, Cheng X, Chen H, et al. Induction of breast cancer stem cells by M1 macrophages through Lin-28B-let-7-HMGA2 axis. Cancer Lett 2019;452:213-25. [Crossref] [PubMed]
- Triboulet R, Pirouz M, Gregory RI. A Single Let-7 MicroRNA Bypasses LIN28-Mediated Repression. Cell Rep 2015;13:260-6. [Crossref] [PubMed]
- Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007;318:1917-20. [Crossref] [PubMed]
- Zhan M, Qu Q, Wang G, et al. Let-7c inhibits NSCLC cell proliferation by targeting HOXA1. Asian Pac J Cancer Prev 2013;14:387-92. [Crossref] [PubMed]
- Zhao B, Han H, Chen J, et al. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett 2014;342:43-51. [Crossref] [PubMed]
- Wang PY, Sun YX, Zhang S, et al. Let-7c inhibits A549 cell proliferation through oncogenic TRIB2 related factors. FEBS Lett 2013;587:2675-81. [Crossref] [PubMed]
- Sun X, Xu C, Tang SC, et al. Let-7c blocks estrogen-activated Wnt signaling in induction of self-renewal of breast cancer stem cells. Cancer Gene Ther 2016;23:83-9. [Crossref] [PubMed]
- Takebe N, Miele L, Harris PJ, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol 2015;12:445-64. [Crossref] [PubMed]
- Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res 2007;13:4042-5. [Crossref] [PubMed]
- Tsanov KM, Pearson DS, Wu Z, et al. LIN28 phosphorylation by MAPK/ERK couples signalling to the post-transcriptional control of pluripotency. Nat Cell Biol 2017;19:60-7. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


