
Study on the interaction mechanism between C-reactive protein and platelets in the development of acute myocardial infarction
Introduction
Acute myocardial infarction (AMI) is a disease in which the heart experiences a sudden deprivation of circulating blood, inducing acute and constant ischemia and hypoxia of the heart and increasing the risk of death (1). Proximately every 40 seconds, someone in the United States experiences a myocardial infarction (MI). AMI accounts for roughly 80% of patients in cardiogenic shock (CS) (2). AMI has two identified subtypes: ST-segment elevation MI (STEMI) and non-STEMI (3). STEMI generally presents as constant ischemic chest pain and elevated serum myocardial necrosis markers, alongside the typical ST-segment elevations on electrocardiogram (ECG) (4). It accounts for approximately 25–40% of AMI (5). The majority of patients with STEMI can receive percutaneous coronary intervention and drugs to relieve symptoms, but it remains an impossible task to rescue the necrotic and apoptotic myocardial cells (6). Unfortunately, no significant decrease has been observed in the risk of death of AMI patients in hospital over the past decades, yet the number of affected patients in China is expected to increase to 23 million by the year 2030 (7). Even if timely treatment exists, acute myocardial ischemic injury and ensuing ischemia-reperfusion injury continue to be challenging issues. Furthermore, radiological and other auxiliary examination approaches, as well as elucidating applicable biomarkers, are regarded as highly necessary for the early diagnosis of this event (8).
Platelets play a key role in thrombotic processes that limit the patency of the recanalized, infarct-related coronary artery and contribute to reperfusion injury (9). The vascular importance of platelets has been attributed to their essential role in mediating MI (10). Studies have shown that platelets become activated during MI (11), but the mechanism of their participation in AMI is not clear. Thus, in our study, we aim to clarify the mechanisms of platelet involvement in the occurrence of AMI through bioinformatics analyses and animal models of AMI in vivo. We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21- 2733).
Methods
Microarray-based analysis concerning up-regulated differentially expressed genes (DEGs)
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
First, the GSE24519 dataset was retrieved from the Gene Expression Omnibus (GEO) database (available at http://www.ncbi.nlm.nih.gov/geo/), which comprised of platform files and original files (CEL files). Next, background correction was implemented, whereupon quantile normalization, prosummarization, and log2 conversion along with a missing values supplement related to the matrix data of GEO were followed with the use of the Robust Multi-array Average algorithm with the R “affy” and “impute” packages. Up-regulated DEGs of GSE24519 were identified and matrices were constructed using the R “limma” package with |log2FC| >1, alongside P value <0.05 working as the threshold.
Weighted gene co-expression network analysis (WGCNA) construction
The GSE24519 dataset included 17 AMI patients with overall survival details, and thus qualified for WGCNA construction. Next, the module eigengenes were linked with the clinical data of AMI patients. A data matrix concerning gene expression in GSE24519 was then constructed, with the first 25% of genes in disease samples exhibiting the largest variance chosen to serve as the input dataset applied for the follow-up WGCNA with the use of the R “WGCNA” package. The outlier sample was then detected and excluded by employing the sample hierarchical clustering method, which was prior to the selection of a qualified soft-threshold power to obtain a standard scale-free network. Thereafter, by means of adjacency, topological overlap matrix (TOM) construction, and corresponding dissimilarity calculation, a gene dendrogram was generated and module identification was carried out with the use of dynamic tree cut, with a minimum module size of 50. Subsequently, module eigengenes cluster analysis was carried out to merge extremely similar modules (with dissimilarity less than 0.25), followed by calculating the correlation of the module eigengene with the AMI clinical phenotype. The module was then selected and used for further analyses, then gene significance (GS) and module membership (MM) were acquired and used for identifying the modules linked to the clinical traits of AMI.
Enrichment analysis
In the gene network in accordance with the scale-free distribution, genes exhibiting identical expression would be modulated together, linked functionally or by shared pathways. Upon topological overlap calculation, genes in the aforementioned chosen module showing a positive association with C-reactive protein (CRP) were investigated. Subsequently, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed via WebGestalt (available at http://www.webgestalt.org/). Functions presenting with P<0.05 were considered statistically significant.
Animals and animal models
Forty purchased specific-pathogen free (SPF) Sprague-Dawley rats (180–220 g, 6–8 W, Hunan, China) were randomized into four groups, namely the AMI group, control group, CRP group, and sham operation group. Prior to the experiment, 2% pentobarbital sodium, sterile equipment packages, and a 5-F bipolar temporary pacing electrode were prepared. The procured rats were food-fasted for a duration of 12 hours, water-fasted for a duration of 6 hours, partially depilated on the back, and weighed. The rats in the AMI group were anesthetized with 2% pentobarbital sodium (0.3 mL/100 g), fixed in the supine position, and intubated with air intake maintained with a ventilator. The leads of the animal ECG monitoring machine were inserted into the subcutaneous muscle layer of limbs according to the operation requirements. After obtaining stable readings on the ECG, second left intercostal thoracotomy was performed to expose the pericardium, and the coronary artery was completely ligated with surgical suture 1–2 mm from the left anterior descending branch. After a few seconds, the ECG showed STEMI, the heart was thereby reset, and the chest was closed layer by layer. The rats in the CRP group were fed a conventional diet and injected with CRP (recombinant human CRP) through the tail vein (3 mg/kg). Meanwhile, the rats randomized into the sham operation group underwent puncture in the left anterior descending branch of the left coronary artery without ligation, and the rest of the process was the same as the model establishment. The rats in the control group were fed a conventional diet without any treatment. Experiments were approved by ethics board of The Second Affiliated Hospital of Qiqihar Medical University, in compliance with Qiqihar Medical University guidelines for the care and use of animals.
Preparation of plasma
Platelet-rich plasma (PRP) is defined as autologous serum with a high concentration of platelets and growth factors, which is obtained by centrifugation of whole blood (11). All rats were anesthetized with 2% pentobarbital sodium at a dose of 0.3 mL/100 g, then blood samples were obtained from the abdominal aorta and collected in a test tube containing 2 mL heparin (5,000 IU/mL) and 800 µL 10% sodium citrate, with an average of 6 mL blood collected per animal. PRP can be prepared via one-step or two-step centrifugation procedures (12). The obtained PRP and platelet-poor plasma (PPP) were prepared and stored at 20 °C. According to the protocol, 10% CaCl2 (10 µL/200 µL PRP) was added to activate PRP before use (13).
Enzyme-linked immunosorbent assay (ELISA)
ELISA is amongst the most significant biochemical analysis techniques for diagnosing and monitoring a plethora of diseases (14). The content of CRP in PPP was detected as per the manufacturer’s instructions of the rat high sensitivity CRP ELISA kit (Sigma, USA).
Real-time quantitative polymerase chain reaction (RT-qPCR)
The total RNA content of PRP, which was isolated using the TRIzol reagent, underwent cDNA reverse transcription with the use of Reverse Transcription kits (PN4366596, acquired from Applied Biosystems). Expression patterns of long non-coding RNAs (lncRNAs) in tissues were quantified using KAPA SYBR FAST qPCR kits (KK4601, acquired from Kapa Biosystems). GAPDH was employed as the internal control, and levels of expressed genes were assessed with the 2–ΔΔCt method combined with MxPro 4.00 (Stratagene), with primer sequences shown in Table 1.

Full table
Statistical analysis
All data involved in this investigation were expressed as mean ± standard deviation (mean ± SD), with a minimum of three repeated independent experiments. Data were analyzed using Graphpad prism 5.0 software. Two-group data comparisons were implemented with Student’s t-test. P<0.05 indicated statistical significance.
Results
Identification of DEGs in AMI
The GSE24519 dataset included 17 AMI patients with detailed gene expression profile information. GSM604591, GSM604585, and GSM604589 were excluded from subsequent analysis in GSE24519 as outlier samples. Therefore, a total of 14 samples with gene expression profile information were included in this study, and 2,852 DEGs (P<0.05, |log2FC| >1) were identified.
Construction of WGCNA and identification of the key module
WGCNA is a well-recognized systematic biological technique often applied for analyzing the expression of genes in varied samples. It is capable of forming either a cluster or module that contains genes presenting with identical expression (15). Once genes are present in a module, they exhibit a tendency to possess similar biological functions, and the associations of modules with sample features can be investigated (16).
The GSM604591, GSM604585, and GSM604589 datasets were removed from follow-up analyses in GSE24519 as outlier samples, leaving 14 samples with survival data that were included in WGCNA (Figure 1A,B). The power of β=6 (scale free R2 =0.9) was chosen as the soft-threshold in order to construct a scale-free network (depicted in Figure 1C,D). Thirteen modules related to gene co-expression were finally identified following removal of the grey module with the use of merged dynamic tree cutting (Figure 1E). A heat map depicting the TOM of the 1000 selected genes showed that each module worked as an independent verification to one another (depicted in Figure 1F). Subsequently, we identified the green-yellow module, representing a key module showing the most significant association with CRP (R2 =0.86, P=7e–05; Figure 1G). Next, we attempted to draw scatter plots concerning GS relative to MM in the green-yellow module with GS for CRP (correlation =0.84, P=3.5e–19; Figure 1H). Gene expression profiles from platelet samples in The Cancer Genome Atlas (TCGA) identified that these 68 genes were over-expressed in AMI samples (Figure 1I).
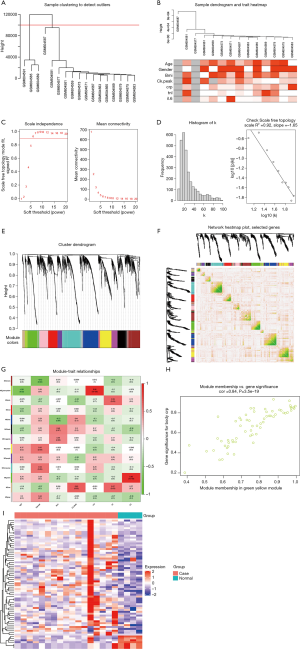
Genes showing a positive association with CRP are uncovered to be enriched in Staphylococcus aureus infection (SAI)
We then investigated the possible biological functions of CRP-related genes in AMI. The results of GO analysis demonstrated that these genes were primarily enriched in biological regulation, metabolic process, cell communication, membrane, nucleus, protein binding, and ion binding, among others (Figure 2A). Further KEGG analysis yielded the correlation between CRP and SAI (Figure 2B).
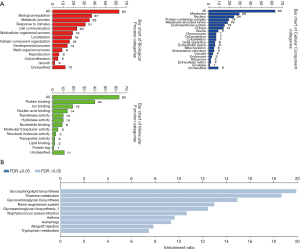
Model diagnosis
The successful establishment of AMI model can be judged by ECG, serum biomarkers or echocardiography, and each method has its important diagnostic significance. In this study, the method of observing the animal signs and ECG in the process of model replication was used as the standard to detect the successful establishment of AMI model. The results of this experiment showed that after ligation of the left anterior descending coronary artery, the myocardial tissue of the left ventricular anterior wall became pale and contraction weakened. Physiological signal acquisition and processing system software system shows that the ECG is converted from normal QRS waveform to typical STEMI. These results indicate that the AMI model was successfully established.
CRP increased in AMI rats
After 6 hours in the animal model, the average levels of CRP in the normal group, sham operation group, AMI group, and CRP group were 1,049±162, 1,674±190, 6,577±836, and 7,949±910 ng/mL, respectively. These findings showed that the content of CRP was increased in the AMI group in contrast to the sham group (**, P<0.01), indicating that AMI can precipitate an increase in CRP. In contrast to the controls, the level of CRP was even higher in the CRP group (**, P<0.01) (Figure 3A,B). Meanwhile, when compared with the normal group, the CRP level in the sham operation group was much higher, which was considered to be related to surgical trauma. However, the increased amplitude and trend were even lower than those in the AMI and CRP groups.
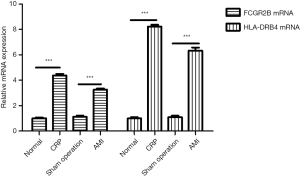
High expression of FCGR2B and HLA-DRB4 links to CRP
In order to verify whether the increase of CRP can influence the expression levels of infection-related genes, the expression patterns of FCGR2B and HLA-DRB4 in the four groups were detected using RT-qPCR. The results illustrated increased mRNA expression levels of FCGR2B and HLA-DRB4 in the AMI group in contrast to the sham operation group (***, P<0.001) (Figure 4). Meanwhile, the mRNA expression levels of FCGR2B and HLA-DRB4 in the CRP group were increased in relation to the control group (***, P<0.001). The increase of CRP may affect the infection-related pathway and aggravate the disease process.
Discussion
AMI, along with heart failure that often ensues, are major causes of morbidity and mortality worldwide (17). Interestingly, the work of our peers has shown that inflammation is key to the pathogenesis of atherosclerosis and acute coronary events (18). Inflammatory responses are a common occurrence after AMI, and can worsen ischemia-reperfusion injuries, triggering augmented infarct size and worse prognoses (19). Moreover, studies have also documented persistently elevated levels of CRP, an inflammation biomarker, in patients following AMI (20).
Firstly, findings obtained in our study from the differential analysis of the GSE24519 dataset using WGCNA revealed a total of 2,852 DEGs. Subsequent findings indicated that the green-yellow module was highly positively correlated with CRP traits. Meanwhile, in vivo experimentation findings demonstrated that the AMI group exhibited significantly higher levels of CRP in contrast to the sham operation group in the STEMI model rats (P<0.001). Elevated levels of CRP have been previously documented in numerous conditions such as renal failure, thyroid disease, and congestive heart failure (21). In our study, GO and KEGG analyses of the 68 genes in the CRP-related green-yellow module in AMI demonstrated that these genes were primarily involved in biological regulation, metabolic process, stimulation response, cell communication, and other biological processes. KEGG pathway analysis illustrated that there were significant differences in three pathways, including SAI, hematopoietic cell lineage, and glycosphingolipid biosynthesis (P<0.05), wherein SAI was found to be the most significant pathway (P=0.0148). The correlation between AMI and infection was first acknowledged during the early 20th century in the context of influenza epidemics. More recently, the association between AMI and SAI has been identified by a case-control and self-control design investigation, which is in accordance with our findings (22).
Inflammatory processes after the advent of MI attract predominant attention in the field of cardiovascular research (23). AMI patients who present with constantly elevated biomarkers related to inflammation (20), in particular CRP, have been reported to be at a greatly increased risk of developing further cardiovascular events (24). CRP is an evolutionarily conserved protein, with homologs present in vertebrates and numerous invertebrates (25). Elevated CRP levels are a common occurrence in inflammatory conditions, cardiovascular diseases, and infections (26). The plasma concentration of CRP was previously shown to deviate by at least 25% in the context of inflammatory disorders (27). Traditionally, CRP works as a non-specific marker related to inflammation. However, research in the last decade has further highlighted the direct involvement of CRP in inflammation and atherosclerosis, as well as other cardiovascular diseases. More importantly, one particular study highlighted CRP as the most powerful predictor and risk factor for cardiovascular disease (28). Furthermore, CRP levels in humans within 6 hours after coronary artery occlusion are known to reflect the inflammatory state of myocardial ischemia (29). The inflammatory reaction is activated within 4–8 hours, and presents with gradual increases in CRP levels (30). In this study, after 6 hours of modeling, CRP in mice increased significantly.
Another notable finding in our study was the increased expression levels of FCGR2B and HLA-DRB4 in addition to CRP in AMI. Inherently, FCGR2B and HLA-DRB4 serve as susceptible genes in autoimmune diseases (31) and play critical roles in maintaining the homeostasis of the immune response (32). FCγRs are well-recognized cell surface receptors that are known to participate in a variety of autoimmune diseases by recognizing the anti-IgG antibody and mediating the immune response, and interacting with the Fc domain of IgG. Currently, there are three different classes of FcγR recognized in humans: FcγRII (CD32), FcγRI (CD64), and FcγRIII (CD16) (33). Platelets adhere and aggregate around bacteria, typically utilizing their GP2b/3a, FcγRIIa, and IgG receptors in conjunction with fibrinogen or fibronectin (34). Various researchers have further documented significant correlations between HLA-DRB and CRP in patients with rheumatoid arthritis, and these genes are even involved in the immune process of myocarditis (35,36). In this study, we established a STEMI rat model in vivo, and assessed the expression patterns of the AMI-related genes FCGR2B and HLA-DRB4 in the SAI pathway using RT-PCR. Our findings showed that the expression levels of FCGR2B mRNA and HLA-DRB4 mRNA in PRP in the AMI group were significantly elevated in contrast to the sham operation group. Compared with the normal group, the expression levels of FCGR2B mRNA and HLA-DRB4 mRNA in PRP in the CRP group were also significantly elevated.
Studies have reported that platelets are involved in the inflammatory response (10). Combined with the results of this study, we speculate that in the development of AMI, the increase in CRP activates platelets and induces platelets to play an anti-inflammatory role.
Acknowledgments
Funding: This study was funded by Qiqihar Science and Technology Program (SFGG-201731).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-2733
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-2733
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-2733). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were approved by ethics board of The Second Affiliated Hospital of Qiqihar Medical University, in compliance with Qiqihar Medical University guidelines for the care and use of animals. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lewis EF, Li Y, Pfeffer MA, et al. Impact of cardiovascular events on change in quality of life and utilities in patients after myocardial infarction: a VALIANT study (valsartan in acute myocardial infarction). JACC Heart Fail 2014;2:159-65. [Crossref] [PubMed]
- Lavall MC, Bagatini MD, Thomé GR, et al. Extracellular hydrolysis of adenine nucleotides and nucleoside adenosine is higher in patients with ST elevation than non-ST elevation in acute myocardial infarction. Clin Lab 2015;61:761-7. [Crossref] [PubMed]
- Kapur NK, Thayer KL, Zweck E. Cardiogenic shock in the setting of acute myocardial infarction. Methodist Debakey Cardiovasc J 2020;16:16-21. [PubMed]
- Gnavi R, Rusciani R, Dalmasso M, et al. Gender, socioeconomic position, revascularization procedures and mortality in patients presenting with STEMI and NSTEMI in the era of primary PCI. Differences or inequities? Int J Cardiol 2014;176:724-30. [Crossref] [PubMed]
- Kook HY, Jeong MH, Oh S, et al. Current trend of acute myocardial infarction in Korea (from the Korea Acute Myocardial Infarction Registry from 2006 to 2013). Am J Cardiol 2014;114:1817-22. [Crossref] [PubMed]
- Kloner RA, Dai W, Hale SL, et al. Approaches to improving cardiac structure and function during and after an acute myocardial infarction: acute and chronic phases. J Cardiovasc Pharmacol Ther 2016;21:363-7. [Crossref] [PubMed]
- Li J, Li X, Wang Q, et al. ST-segment elevation myocardial infarction in China from 2001 to 2011 (the China PEACE-Retrospective Acute Myocardial Infarction Study): a retrospective analysis of hospital data. Lancet 2015;385:441-51. Retracted and republished in Lancet 2015;385:402. [Crossref] [PubMed]
- Ye XD, He Y, Wang S, et al. Heart-type fatty acid binding protein (H-FABP) as a biomarker for acute myocardial injury and long-term post-ischemic prognosis. Acta Pharmacol Sin 2018;39:1155-63. [Crossref] [PubMed]
- Gawaz M, Neumann FJ, Ott I, et al. Platelet function in acute myocardial infarction treated with direct angioplasty. Circulation 1996;93:229-37. [Crossref] [PubMed]
- Koupenova M, Clancy L, Corkrey HA, et al. Circulating platelets as mediators of immunity, inflammation, and thrombosis. Circ Res 2018;122:337-51. [Crossref] [PubMed]
- Xiao SJ, Zhou YF, Wu Q, et al. Uncovering the differentially expressed genes and pathways involved in the progression of stable coronary artery disease to acute myocardial infarction using bioinformatics analysis. Eur Rev Med Pharmacol Sci 2021;25:301-2. [PubMed]
- Yazigi JA Junior, Dos Santos JB, Xavier BR, et al. Quantification of platelets obtained by different centrifugation protocols in SHR rats. Rev Bras Ortop 2015;50:729-38. [PubMed]
- Xu J, Gou L, Zhang P, et al. Platelet-rich plasma and regenerative dentistry. Aust Dent J 2020;65:131-42. [Crossref] [PubMed]
- Zhdanov A, Keefe J, Franco-Waite L, et al. Mobile phone based ELISA (MELISA). Biosens Bioelectron 2018;103:138-42. [Crossref] [PubMed]
- Beckerman P, Qiu C, Park J, et al. Human kidney tubule-specific gene expression based dissection of chronic kidney disease traits. EBioMedicine 2017;24:267-76. [Crossref] [PubMed]
- Tang J, Kong D, Cui Q, et al. Prognostic genes of breast cancer identified by gene co-expression network analysis. Front Oncol 2018;8:374. [Crossref] [PubMed]
- Ong SB, Hernández-Reséndiz S, Crespo-Avilan GE, et al. Inflammation following acute myocardial infarction: multiple players, dynamic roles, and novel therapeutic opportunities. Pharmacol Ther 2018;186:73-87. [Crossref] [PubMed]
- Newby LK. Inflammation as a treatment target after acute myocardial infarction. N Engl J Med 2019;381:2562-3. [Crossref] [PubMed]
- Akodad M, Sicard P, Fauconnier J, et al. Colchicine and myocardial infarction: a review. Arch Cardiovasc Dis 2020;113:652-9. [Crossref] [PubMed]
- Thackeray JT, Hupe HC, Wang Y, et al. Myocardial inflammation predicts remodeling and neuroinflammation after myocardial infarction. J Am Coll Cardiol 2018;71:263-75. [Crossref] [PubMed]
- Liu Z, Ma C, Gu J, et al. Potential biomarkers of acute myocardial infarction based on weighted gene co-expression network analysis. Biomed Eng Online 2019;18:9. [Crossref] [PubMed]
- McNamara JF, Harris PNA, Chatfield MD, et al. Acute Myocardial Infarction and Community-Acquired Staphylococcus aureus Bloodstream Infection: an observational cohort study. Clin Infect Dis 2020; Epub ahead of print. [Crossref] [PubMed]
- Rischpler C. Acute myocardial infarction. Q J Nucl Med Mol Imaging 2016;60:236-51. [PubMed]
- Hennessy T, Soh L, Bowman M, et al. The Low Dose Colchicine after Myocardial Infarction (LoDoCo-MI) study: a pilot randomized placebo controlled trial of colchicine following acute myocardial infarction. Am Heart J 2019;215:62-9. [Crossref] [PubMed]
- Black S, Kushner I, Samols D. C-reactive protein. J Biol Chem 2004;279:48487-90. [Crossref] [PubMed]
- Du Clos TW, Mold C. C-reactive protein: an activator of innate immunity and a modulator of adaptive immunity. Immunol Res 2004;30:261-77. [Crossref] [PubMed]
- Sproston NR, Ashworth JJ. Role of C-reactive protein at sites of inflammation and infection. Front Immunol 2018;9:754. [Crossref] [PubMed]
- Osimo EF, Baxter LJ, Lewis G, et al. Prevalence of low-grade inflammation in depression: a systematic review and meta-analysis of CRP levels. Psychol Med 2019;49:1958-70. [Crossref] [PubMed]
- Geluk CA, Post WJ, Hillege HL, et al. C-reactive protein and angiographic characteristics of stable and unstable coronary artery disease: data from the prospective PREVEND cohort. Atherosclerosis 2008;196:372-82. [Crossref] [PubMed]
- Anzai T. Inflammatory mechanisms of cardiovascular remodeling. Circ J 2018;82:629-35. [Crossref] [PubMed]
- Yuksel M, Xiao X, Tai N, et al. The induction of autoimmune hepatitis in the human leucocyte antigen-DR4 non-obese diabetic mice autoimmune hepatitis mouse model. Clin Exp Immunol 2016;186:164-76. [Crossref] [PubMed]
- Su K, Wu J, Edberg JC, et al. A promoter haplotype of the immunoreceptor tyrosine-based inhibitory motif-bearing FcgammaRIIb alters receptor expression and associates with autoimmunity. I. Regulatory FCGR2B polymorphisms and their association with systemic lupus erythematosus. J Immunol 2004;172:7186-91. [Crossref] [PubMed]
- Vogelpoel LT, Baeten DL, de Jong EC, et al. Control of cytokine production by human fc gamma receptors: implications for pathogen defense and autoimmunity. Front Immunol 2015;6:79. [Crossref] [PubMed]
- Loughman A, Fitzgerald JR, Brennan MP, et al. Roles for fibrinogen, immunoglobulin and complement in platelet activation promoted by Staphylococcus aureus clumping factor A. Mol Microbiol 2005;57:804-18. [Crossref] [PubMed]
- Klimenta B, Nefic H, Prodanovic N, et al. Association of biomarkers of inflammation and HLA-DRB1 gene locus with risk of developing rheumatoid arthritis in females. Rheumatol Int 2019;39:2147-57. [Crossref] [PubMed]
- Şelli ME, Thomas AC, Wraith DC, et al. A humanized HLA-DR4 mouse model for autoimmune myocarditis. J Mol Cell Cardiol 2017;107:22-6. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


